Abstract
Crop improvement, conservation and utilization of plant genetic resources has benefited from chromosomal studies of germplasms. In the Indian sub continent, lentil (Lens culinaris Medik.) is a major rabi pulse crop with 2n = 2x = 14 chromosomes. Chromosomal studies were carried out on Indian lentils during the period of 1952 to 1991 employing orcein squash techniques by different authors but there was no unanimity on chromosome morphology and chromatin length. This is the first study conducted on two wild and 12 cultivated cultivars of Lens for detailed and comparative chromosomal analysis using enzymatic maceration and air drying (EMA) based Giemsa staining methods. Chromosomal analysis revealed chromosomal stability, uniform karyotype formula (3m + 1m (sat) + 2sm + 1st), one pair of interstitial sat in either chromosome number 3 or 4 and interesting variations in total chromatin length (53.6–121.2 μm) in all the studied cultivars. More studies on other species and cultivars along with the application of flurochrome dyes for characterizing apparently similar chromosomes at the cultivar level may be very useful for future crop improvements.
Keywords:
Introduction
In the Indian sub continent, lentil (Lens culinaris Medik) is a major rabi pulse crop and due to its high protein content it is referred to as “poor man’s meat”. It contains high bio-available minerals, beta-carotene, dietary fiber, foliate, and low phytates (Thavarajah and Thavarajah Citation2011). The genus Lens is known as masoor or malaka and belongs to the family Fabaceae (Leguminosae), subfamily Faboideae, tribe Fabeae. The genus comprises one cultivated and six wild species (van Oss et al. Citation1997). Lens culinaris Medikus, the only cultivated species, is reported to have originated from a wild species Lens orientalis. Lentil is an herbaceous self-pollinated annual plant, with a low out-crossing frequency. The cultivated species Lens culinaris subsp. culinaris is divided into two groups: the small seeded, red cotyledon var. microsperma, and the large seeded, green var. macrosperma. The taxonomy of the genus is still open to discussion given the close relationships among its species. India is the largest importer, second highest producer, processor and consumer of lentils and encourages different cultivars for large-scale cultivation. Due to its high domestic consumption there remains a gap between demand and supply. Climatic, biotic and abiotic stresses, use of poor and marginal soil and continuous dependency on synthetic fertilizers, insecticides and pesticides are the main constraints in genetic improvements of lentil. Crop improvement, conservation and utilization of plant genetic resources has benefited from chromosomal studies of germplasms. Chromosome analysis can reveal whether the plant is stable at the chromosomal level and whether additional variation originates over a long period of time against climatic, biotic and abiotic stresses. A literature review revealed that all the species, sub-species, cultivars, and germplasms of Lens contain 2n = 2x = 14 chromosomes. There have been a number of chromosomal studies using ocean squash techniques on selected cultivars of Indian lentils, but there was no unanimity on chromosome morphology (meta centric, sub metacentric and acrocentric), karyotype formula or total chromatin length (Bhattacharjee Citation1951; Sharma and Mukhopadhyaya Citation1963; Sinha and Acharia Citation1972; Naithani and Sarbhoy Citation1973; Gupta and Singh Citation1981; Lavania and Lavania Citation1983; Nandanwar and Narkhede Citation1991).
Ladizinsky (Citation1993) used Feulgen staining and observed that the Lens karyotype contains three types of chromosome (metacentric or submetacentric, metacentric with a sat and acrocentric chromosomes) and formulated 3 m/sm + 1m with sat + 3ac as their karyotypic formula. Balyan et al. (Citation2002) also used Feulgen staining as well as fluorescent in situ hybridization (FISH) for his chromosomal analysis and observed that the position of secondary constriction in cultivated and some wild species, namely L. culinaris, L. orientalis, L. odemensis and L. ervoides, was close to the centromere, i.e. interstitial. However in L. nigricans the position of secondary constriction was near the telomere of an acrocentric chromosome pair. Galasso (Citation2003) conducted FISH to study the repeated DNA sequences on different species of Lens, and Fernández et al. (Citation2005) analyzed rDNA genome regions in some Lens species using non transcribed spacer (NTS) and FISH.
A chromosomal reassessment of a large number of lentil cultivars in India was felt to be necessary. It also appears that no Indian cultivars have been analyzed using the enzymatic maceration and air drying (EMA) method. This method was first demonstrated by Kurata and Omura (Citation1978) for analysis of plant chromosomes and is useful for preparing chromosomes from plants with small chromosomes (Fukui Citation1996). The EMA method followed by Giemsa can provide chromosomes in a cytoplasm-free clear background with distinct chromosome morphology. Precise karyotype analysis as well as molecular cytogenetic analysis is possible with the EMA slides. We have been able to collect many wild species and widely cultivated cultivars of Lens culinaris from the Indian Institute of Pulses, Kanpur, India. Thus the aim of the present study was to standardize EMA based preparation of chromosomes from 14 Lens cultivars and their comparative analysis following Giemsa staining.
Materials and methods
Fourteen Indian germplasms from the cultivated and wild species of L. culinaris, L. orientali and L. odemensis were employed for detailed karyotypic analysis. Except for three local cultivars all the samples were obtained from the Indian Institute of Pulses, Kanpur, India. For each cultivar 25–30 seeds, and five seeds from each wild species, were germinated in dark on moist filter paper for about two days for harvesting root apical meristems. Roots from all the samples were collected, pre-treated with a saturated solution of para-dichlorobenzene at 16°C for 4–5 h, fixed at 3:1 (methanol and acetic acid) solution for more than two hours and finally stored at –20°C for all chromosomal analyses. Fixed roots were treated with cold water at 4°C for 3–12 h and then placed in a cocktail enzyme mixtures (cellulose Onozuka RS 1%, macerozyme R-10, 0.75%, pectolyase Y-23 0.15% and 1 mM EDTA) for 45–55 minutes at 37°C. After washing the enzyme-treated roots with water they were teased uniformly on cleaned slides with 1:3 acetic methanol and air dried. Completely air dried slides were dipped in 2% Giemsa solution (Merck, Darmstadt, Germany) in 1/15 phosphate buffer solution for 10–14 min at room temperature, washed thoroughly with doubled distilled water and air dried for chromosomal analysis. A few slides were also prepared using orcein squash method following the standard protocol of Sharma and Sharma (Citation1980). Chromosomal documentation and analysis was carried out using a Carl Zeiss AxioLab A1 microscope (Jena, Germany), fitted with a CCD camera, computer and Axiovision L.E 4 software. Photographs of a minimum of five Giemsa stained metaphase plates were used to measure and calculate different chromosomal parameters [length of long (l) and short (s) arms; absolute chromatin length; total diploid chromatin length; arm ratios (l/s) to determine centromeric position as per Levan et al. (Citation1964); centromeric index (short arm length/total chromatin length × 100); and relative length (absolute chromatin length/total diploid chromatin length)]. Karyomorphometric data were used to prepare idiograms and karyotypic formula for individual cultivars. Statistical analysis was performed as per our earlier design (Dafadar et al. Citation2012).
Results and discussion
Variation in germination percentage was noted among the cultivars. While a maximum of 97.14% germination was obtained in a microtype cultivar from Bankura, the lowest (36.36%) was from a cultivar named KLS210 (Table ). To determine chromosome number and numerical variations, three to five roots per cultivar and a minimum of 25 countable metaphase plates were studied. Chromosomal analysis in all the species and cultivars under studies was primarily carried out through EMA method followed by Giemsa staining. However, orcein squash in addition to Giemsa was conducted in two L. culinaris cultivars, namely macro and micro Barasat (Table M, N) to compare chromosome morphology. Using the EMA method we have been able to score a large number of very well scattered metaphase plates in all the species and cultivars of Lens. Metaphase plates are free from cytoplasmic background and with distinct chromosome morphology. Identification of primary and secondary constriction regions is not difficult (Figures –). On the other hand orcein squash results in very few metaphase plates (Figure I) without cytoplasmic background and with distinct primary and with secondary constriction. Moreover, EMA preparations but not orcein squash are useful in molecular cytogenetic studies.
Table 1. List of the Lens accessions studied with their source and germination efficiency.
Figure 1. Giemsa stained well-scattered somatic metaphase plates of nine Lens species and cultivars (2n =14): (A) L. odemensis (ILWL 35); (B) L. orientalis (ILWL 248); (C) L. culinaris (IPL81); (D) L. culinaris (IPL316); (E) L. culinaris (JL-1); (F) L. culinaris (Pl406); (G) L. culinaris (KLS 210); (H) L. culinaris (EC704030); (I) L. culinaris (EC78455). Scale bars: 5 μm.
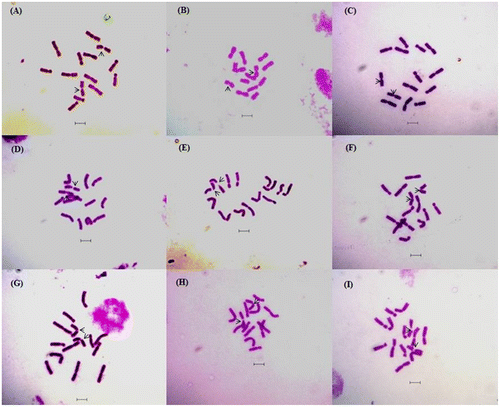
Figure 2. Giemsa and orcein stained well-scattered somatic metaphase plates in different cultivars of L. culinaris: (A) L. culinaris (EC 78498); (B) L. culinaris (EC267877); (C) L. culinaris (micro, Bankura); (D) L. culinaris (macro, Barasat); (E) L. culinaris (micro, Barasat); (F) monosomic metaphase plate of L. culinaris (EC, 78410) showing 2n = 13 chromosomes; (G) monosomic metaphase plate of L. culinaris (IPL81) showing 2n = 26 almost separated chromatids; (H) L. culinaris ((IPL316) metaphase plate showing 2n = 28 almost separated chromatids; (I) orcein stained somatic metaphase plate with 2n = 14 chromosomes in L. culinaris (macro Barasat). Scale bars: 5 μm.
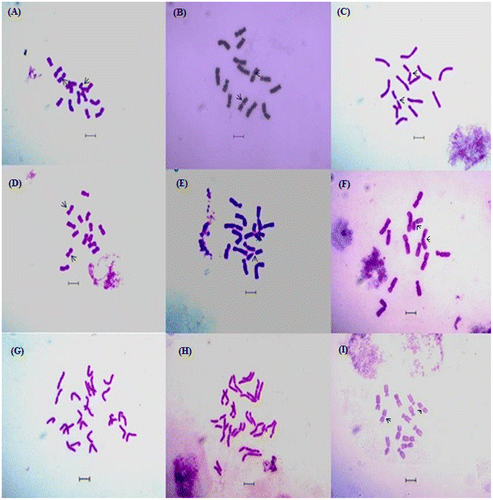
Chromosomal analysis confirms 2n = 2x = 14 large type chromosomes (Fukui Citation1996) in all lentil species and cultivars (Figures A-I, J). No significant numerical variations were observed, except in some monosomic cells (2n – 1) with 2n = 13 chromosomes in cultivars IPL 81, Macro Barasat, EC 78410 (NBPGR), and EC 78498(NBPGR) (Figure F, G). The results indicate chromosomal stability in all the cultivars of lentils obtained from different climatic sources and growing against all biotic and abiotic stresses. In one exception, a trisomic cultivar from Bangladesh (Khandaker et al. Citation2007), chromosomal stability was reported in different species of Lens (Balyan et al. Citation2002). In most of the previous reports from India and Iran extreme variation was reported in average chromosome length and total diploid chromatin length within the cultivars of L. culinaris.
Total chromatin length of any cultivar is related to the length of the individual chromosomes within a genome. The average length of the individual chromosome has been found as 3.8–9.8 μm (Sharma and Mukhopadhyaya Citation1963); 3.05 μm (Naithani and Sarbhoy Citation1973); 4.0–7.36 μm (Gupta and Singh Citation1981); 4.54 μm (Dixit and Dubey Citation1985); 1.9–4.3 μm (Lavania and Lavania Citation1983); and 3.26 μm (Rehman and Altaf Citation1994). Average chromosome length in all the 14 cultivars analyzed in our studies ranged from 3.2 to 10.4 μm (Table ), which is nearly identical to the observation obtained five decades ago by Sharma and Mukhopadhyaya (Citation1963). Gupta and Singh (Citation1981) obtained 39.3 μm diploid TCL in the variety Pant L639. Dixit and Dubey (Citation1986) reported variation of 28.2–72.3 μm in different microsperma cultivars of L. culinaris cul. culinaris and the lowest chromatin length of 16.9 μm was reported by Lavania and Lavania (Citation1983). On the other hand our results on total chromatin length from 12 Indian cultivars of L. culinaris as well as two wild species, L. odemensis and L. orientalis, revealed a range from 53.6 to 121.2 μm (Table ). Balyan et al. (Citation2002) reported 25.87 μm diploid chromatin lengths in L. odemensis, whereas we have obtained 94.8 μm in the same wild species.
Table 2. Comparative karyometric analysis of 14 Lens species and cultivars.
We have recorded more than 100 μm chromatin length in seven cultivars (Table ), not reported earlier in any Indian Lens species or cultivar. It is obvious that variation of chromosome length will lead to variation in total diploid chromatin length. But it is difficult to explain the reasons for such types of variation. It is assumed that methods adopted for chromosome processing, the exact position of root meristematic cells within a root, and physiological differences between the roots at the time of processing may exert some influence on such types of variation. Results of experimental variation may lead to intraspecific C-value variation. C-value variation can also be correlated with eco-geographic variables, suggesting that the variation is adaptive and that these may be examples of incipient speciation (Murray Citation2005). We have adopted uniform methods of chromosome preparation for all the 14 cultivars and thus negate the first assumption of experimental variation. Leitch and Leitch (Citation2013) considered amplification of non-coding repetitive DNA, especially retrotransposons, as one possible mechanism responsible for generating changes in genome size in different land plant groups. It is important to note that genome size variations have not changed the diploid chromosome numbers in Lens species and cultivars over a long period of time.
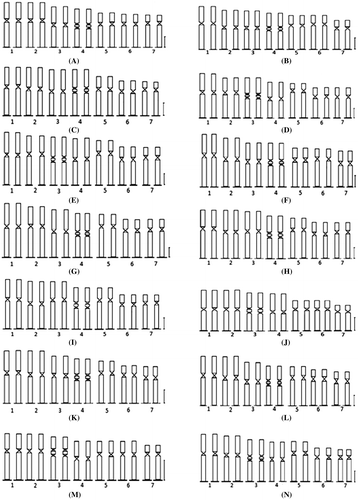
There was another controversy related with the presence/absence and number of sat chromosomes in Lens karyotype. Naithani and Sarbhoy (Citation1973) reported two pairs of sat chromosomes in L. culinaris var macro and microsperma. Bhattacharjee (Citation1951), Sharma and Mukhopadhyaya (Citation1963), Sinha and Acharia (Citation1972), Ladizinsky (Citation1993), and Gaffarzadeh-Namazi et al. (Citation2007) had reported one pair interstitial sat chromosome in Lens. Sinha and Acharia (Citation1972) reported absence of sat chromosome in Russian and EC verities of L. culinaris. Difficulties exist in locating the position and number of sat chromosome in Lens if it is not handled carefully and this may be the reason for citing 0–3 sat chromosomes. However, adopting EMA based chromosome preparation, we have been able to obtain cytoplasm-free well-scattered metaphase plates in all the cultivars and identified a pair of interstitial sat chromosomes in all the 14 cultivars (Figure A–N). The next question was: which chromosome pairs contain secondary constriction or sat in Lens? It was reported that the secondary constriction was present on the fifth chromosome in L. ervoides and on the fourth chromosome in other species (Gupta and Singh Citation1981; Gaffarzadeh-Namazi et al Citation2007). Our digital documentation and hand drawings (Figures –) of chromosome morphology revealed that the sat chromosome may be present not only in chromosome 4 but in some cases in chromosome 3. We have prepared idiograms and karyotype formulae (Figure , Table ). In Lens, Sindhu et al. (Citation1983) reported the presence of metacentric, submetacentric and subterminal chromosomes [3m (1 sat included) + 1sm + 3st] in three species, namely L. culinaris, L. orientalis and L. nigricans, and considered this as a general karyotype formula. Gupta and Singh (Citation1981) reported metacentric and submetacentric chromosomes in the karyotype formula (4 m + 3 sm) in L. culinaris variety Pant L 639. Lavania and Lavania (Citation1983) suggested karyotype formula with one type of chromosomes (7sm) in a lens variety 3847. Ladizinsky (Citation1993) presented either metacentric or submetacentric and acrocentric chromosomes in Lens karyotype (3m or 3sm + 1m (sat) + 3ac). Gaffarzadeh-Namazi et al. (Citation2007) reported a metacentric and submetacentric chromosome formula [3m + 1m (sat) + 3sm] for 10 landraces of L. culinaris of Iran. Variation in karyotypic formula (Levan et al. Citation1964) within and between the species is not uncommon. In our chromosomal analysis we have prepared the karyotype formula for 14 Indian cultivars of Lens, and found homology with Sindhu et al. (Citation1983) regarding the category of chromosomes present in the karyotypic formula. However, some differences exist with the number of chromosomes under each category. We have obtained metacentric, submetacentric, subterminal and sat chromosomes in all our cultivars and thus could not agree with many of the above noted authors. The results of chromosomal analysis in the present study could not find any variation in karyotype formula within the 14 cultivars of Lens and present a uniform formula of 3m + 1m (sat) + 2 sm + 1st. We have obtained identical karyotype formulae between L. culinaris and L. orientalis, the wild progenitor of the former. A similar observation was reported by Ladizinsky (Citation1979). Variation in total chromatin length exists among the Indian cultivars (Table ). Exchange of chromosomal segments between nearly similar sized chromosomes during the evolution of the genus is one possibility. Chromosome interchanges between different species of Lens has been reported by Tadmor et al. (Citation1987) and Balyan et al. (Citation2002) and our results support the above opinion.
Figure 3. Hand drawings of Lens species and cultivars showing position of one pair interstitial sat chromosomes: (A) L. odemensis (ILWL 35); (B) L. orientalis (ILWL 248); (C) L. culinaris (IPL81); (D) L. culinaris (IPL316); (E) L. culinaris (JL-1); (F) L. culinaris (Pl406); (G) L. culinaris (KLS 210); (H) L. culinaris (EC704030); (I) L. culinaris (EC78455); (J) L. culinaris (EC 78498); (K) L. culinaris (EC267877); (L) L. culinaris (micro, Bankura); (M) L. culinaris (macro, Barasat); (N) L. culinaris (micro, Barasat). Scale bars: 5 μm.
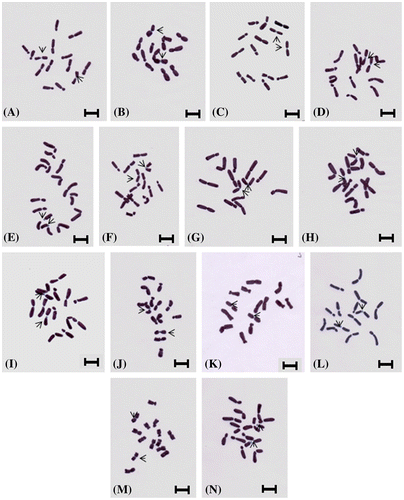
Figure 4. Idiograms of 14 species and cultivars of Lens. Chromosomes are arranged in order of descending length: (A) L. odemensis (ILWL 35); (B) L. orientalis (ILWL 248); (C) L. culinaris (IPL81); (D) L. culinaris (IPL316); (E) L. culinaris (JL-1); (F) L. culinaris (Pl406); (G) L. culinaris (KLS 210); (H) L. culinaris (EC704030); (I) L. culinaris (EC78455); (J) L. culinaris (EC 78498); (K) L. culinaris (K. EC2678); (L) L. culinaris (micro, Bankura); (M) L. culinaris (macro, Barasat); (N) L. culinaris (micro, Barasat). Scale bars: 2 μm.
The present study has presented the karyotypic analysis of two wild and 12 cultivated cultivars of Lens, applying the EMA method of chromosome preparation, and presents a new chromosomal database. Chromosomal analysis revealed numerical stability, uniform karyotype formulae, presence of one pair of interstitial sat chromosomes in chromosome number 3 or 4 and, more interestingly, variations in total chromatin length in all the studied cultivars. However, more studies are required in other species and cultivars using EMA methods. Application of fluorescent banding and FISH in future may help to characterize apparently similar chromosomes at the constitutive heterochromatin level as an aid for future crop improvements.
Acknowledgements
TBJ acknowledges UGC for awarding the project and Director and Dr. J. Kumar of Indian Institute of Pulses, Kanpur, India along with the Principal Barasat Govt. College and Sayantani Nath for providing the plant materials, basic facilities and computational work. TBJ dedicates the paper to Prof. A.K. Sharma and Prof. S. Sen of the University of Calcutta.
Disclosure statement
There is no conflict of interest.
Additional information
Funding
References
- Balyan HS, Houben A, Ahne R. 2002. Karyotype analysis and physical mapping of 18S-5.8S-25S and 5S ribosomal RNA loci in species of genus Lens Miller (Fabaceae). Caryologia. 55(2):121–128.
- Bhattacharjee SK. 1951. Cytogenetics of Lens esculenta Moench Microsperma. Sci Cult. 16:426–427.
- Dafadar A, Das A, Bandyopadhyay S, Jha TB. 2012. In vitro propagation and molecular evaluation of a Capsicum annuum L. cultivar with a high chromosome number (2n = 48). Sci Hortic. 140:119–124.
- Dixit P, Dubey DK. 1985. Heritability and genetic advance in induced mutants of lentil (Lens culinaris Medik.). Ind J Genet. Plant Breed. 45(3):520–524.
- Dixit P, Dubey DK. 1986. Karyotype study in Lentil. Lens newsletter 13(1): 8–10.
- Fernández M, Ruiz ML, Linares C, Fominaya A, Pérez de la Vega M. 2005. the 5S rDNA genome regions of Lens species. Genome. 48(5):937–942.
- Fukui K. 1996. Plant chromosome at mitosis. In: Fukui K, Nakayama S, editors. Plant Chromosome Laboratory Methods. Boca Raton: CRC Press; p. 1–17.
- Gaffarzadeh-Namazi L, Asghari-Zakaria R, Babaeian N, Kazemi-Taber K. 2007. Comparative study of chromosome morphology and C-banding patterns in several genotypes of Lens culinaris. Pak J Biol Sci. 10(11):1811–1816.
- Galasso I. 2003. Distribution of highly repeated DNA sequences in species of the genus Lens Miller. Genome. 46(6):1118–1124.
- Gupta PK, Singh J. 1981. Standard karyotype in lentil (Lens culinaris Med.) var. PL-639. LENS. 8:23.
- Khandaker M, Imdadul Hoque M, Alam SS. 2007. Fluorescent banding in three varieties of Lens culinaria Medik (Fabaceae). Cytologia. 72(2):227–231.
- Kurata N, Omura T. 1978. Karyotype analysis in rice, 1: A new method for identifying all chromosome pairs. Japan J Genet. 53(4):251–255.
- Ladizinsky G. 1979. The origin of lentil and its wild gene pool. Euphytica. 28:179–187.
- Ladizinsky G. 1993. Wild lentils. Crit Rev. Plant Sci. 12:169–184.
- Lavania UC, Lavania S. 1983. Karyotype studies in Indian pulses. Genet Agrar. 37:299–308.
- Leitch LJ, Leitch AR. 2013. Genome size diversity and evolution in land plants. In: Leitch IJ, Greilhuber J, Dolezel J, Wendel JF., editors. Plant genome diversity (Vol. 2). Wien: Springer-Verlag. p. 307–320.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromere position on chromosomes. Hereditas. 52:201–220.
- Murray BG. 2005. When does intraspecific C-value variation become taxonomically significant? Ann Bot. 95:119–125.
- Naithani SP, Sarbhoy RK. 1973. Cytological studies in Lens esculenta Moench. Cytologia. 38:195–203.
- Nandanwar RS, Narkhede MM. 1991. Intraspecific variation in karyotype of lentil. J Maharastra Agri Univ. 16:24–27.
- Rehman S, Altaf CM. 1994. Karyotypic studies in Lens culinaris Medic, S.Sp Macrosperma cv. Laird x Precoz. Pak J Bot. 26(2):347–352.
- Sharma AK, Mukhopadhyaya S. 1963. Karyotype constancy in different strains of Lens esculentum Moench as worked out through recent techniques. Ind Agric. 7:103–111.
- Sharma AK, Sharma A. 1980. Chromosome techniques. Theory and practice (3rd ed.) London: Butterworth’s.
- Sindhu JS, Slinkard AE, Scoles GJ. 1983. Studies on variation in Lens I karyotype. Lens. 10:14.
- Sinha SS, Acharia SS. 1972. Karyotype analysis in some varieties of Lens culinaris. Cytologia. 37:673–683.
- Tadmor Y, Zamir D, Ladizinsky G. 1987. Genetic mapping of an ancient translocation in the genus Lens. Theor Appl Genet. 73:883–892.
- Thavarajah D, Thavarajah P. 2011. Lentil (Lens culinaris) as a biofortified crop with essential micronutrients: A food-based solution to micronutrient malnutrition. Grain Legume. 57:29–31.
- van Oss H, Aron Y, Ladizinsky G. 1997. Chloroplast DNA variation and evolution of the genus Lens Mill. Theor Appl Genet. 94:452–457.
- Wang L, An L, Hu Y, Wei L, Li Y. 2009. Influence of phytohormones and medium on the shoot regeneration from leaf of Swertia chirata Buch.-Ham. Ex wall. in vitro. Afr J Biotech. 8:2513–2517.
