Abstract
The number and behavior of meiotic chromosomes in four populations of three species of Vernonanthura were analyzed. It was observed that all stages of meiosis were stable without chromosomal irregularities. The somatic chromosome number is reported for four populations of three species of Vernonanthura and two populations of Vernonia. All species were diploid with 2n = 34. The first chromosome count is presented for Vernonanthura chaquensis (2n = 17II and 2n = 34). The karyotypes were determined for Vernonanthura lucida (32m + 2sm), V. oligactoides (24m + 10sm) and V. pseudolinearifolia (28m + 6sm); they are presented for the first time. The total length of the karyotype was 31.67μ–40.48 μm. The average chromosome length was 1.86μ–2.38 μm and the centromeric index was 38.61–42.99. Vernonanthura oligactoides showed the smallest variation of chromosome length within the karyotype (1.42 μm) while V. squamulosa had the biggest (2.15 μm). The different karyotypic parameters were analyzed with two statistic tests. The resulting species arrangement from Unweighted Pair Group Method with Arithmetic Mean (UPGMA) grouping analysis fully fits with that obtained with principal component analysis (PCA). Both tests demonstrated congruence between karyotypic parameter and classification based on morphological characters.
Introduction
The Vernonieae Cass. (Asteraceae) are a pantropical tribe of 89 genera which are concentrated around two major centers of diversification, the central region of Africa and southern Brazil. Traditional classification grouped the members of the tribe into six different subtribes based on inflorescence patterns, persistence of phyllaries, pollen morphology, chemical composition and chromosome numbers (Bremer Citation1994). The subtribe Vernoniinae Less constitutes the largest group within the Vernonieae, including approximately 1100 species. This group comprises almost all the species previously placed into the huge genus Vernonia Schreb.
Morphological and molecular phylogenetic studies had led to the recognition of 21 subtribes, 15 in the New World and six in the Old World. The new classification proposed significant taxonomic changes, resulting in the reduction in the size and distribution of the core genus Vernonia and increment of the new genera. Now, there are 125 genera, 50 of them are monotypic and 35 with fewer than three species (Keeley and Robinson Citation2009). Chromosome numbers have not been utilized in the proposed classification and less than 20% of the species from the New World have been analyzed (Dematteis Citation2002). However, the cytological data from South America Vernonieae show that chromosomes are widely variable in numbers and morphology, which suggest that can be useful in the taxonomic and evolutionary studies (Ruas et al. Citation1991; Dematteis and Fernández Citation1998, Citation2000; Angulo and Dematteis Citation2009a; Via do Pico and Dematteis Citation2012).
The genus Vernonanthura H. Rob. was established to separate the early taxa arranged under Vernonia sect. Lepidaploa Paniculatae Benth. and Hook., which consists of about 80 species distributed from southern Mexico to the central Argentina, but mostly concentrated in southeastern Brazil (Robinson Citation1992, Citation1995; Vega and Dematteis Citation2011a). This genus is closely related to Vernonia s. str. because they both show the same type of pollen and basic chromosome number (Vega and Dematteis Citation2011b, Citation2012), but differ in the inflorescence type, habitat and sometimes by the presence of tailed anther bases (Robinson Citation1992). The members of the new genus are shrubs or small trees having thyrsoid to pyramidal inflorescences, with individual branches cymose to corymbiform (Robinson Citation1992). In southern South America, the species are mainly concentrated in the mountains of northwest Argentina and the fields and forests of Paraguay and eastern Argentina (Cristóbal and Dematteis Citation2003; Vega and Dematteis Citation2008).
Almost all the cytological studies of Vernonanthura species were carried out under the old concept of Vernonia (Hunter Citation1964; Jones Citation1979, Citation1982; Galiano and Hunziker Citation1987; Stutts Citation1988; Ruas et al. Citation1991; Dematteis Citation1996, Citation1997, Citation2002; Dematteis and Fernández Citation1998; Salles de Melo et al. Citation2010; Oliveira et al. Citation2007). The single analysis after the genus segregation reported chromosome numbers of 15 populations belonging to 10 taxa of Vernonanthura (Vega and Dematteis Citation2012). In this study, first chromosome counts were presented for three taxa: V. lucida (n = 17II, 2n = 34); V. oligactoides (n = 17II, 2n = 34) and V. pseudolinearifolia (2n = 34). The genus Vernonanthura seems to be cytologically homogeneous with basic number x = 17 and almost all diploid taxa with 2n = 34.
The genus Vernonia sens. str. has only two South American species, V. incana, which is always diploid with 2n = 2x = 34, and V. echioides with diploid and tetraploid cytotypes (Vega and Dematteis Citation2012).
The karyotype of seven species of Vernonia sensu lato were analyzed by Dematteis and Fernández (Citation1998), six of them now included in Vernonanthura. These taxa show rather similar chromosomal features, although some differences in karyotype formula, chromosome size, and symmetry.
The chromosome number of about 30% of the species of Vernonanthura are known and the karyotypes of about 10% of these were analyzed (Hunter Citation1964; Jones Citation1974, Citation1979, Citation1982; Stutts Citation1988; Galiano and Hunziker Citation1987; Dematteis Citation1996, Citation1997, Citation2002; Dematteis and Fernández Citation1998; Carr et al. Citation1999; vCitation2007; Salles de Melo et al. Citation2010; Vega and Dematteis Citation2012).
The purpose of this study was to determine the chromosome number and karyotype formula of some species of Vernonia and Vernonanthura. The cytological data are discussed in relation to the taxonomy and the chromosome evolution of both genera.
Material and methods
The populations studied were collected in the field. Voucher specimens are deposited in the herbarium of Instituto de Botánica del Nordeste (CTES). The sources of examined species and somatic numbers are presented in Table .
Table 1. Specimens examined, meiotic number (n) and mitotic number (2n) observed in a population of Vernonanthura and two populations of Vernonia incana.
The meiotic analyses were carried out from flower buds previously fixed in ethanol and lactic acid in the proportion of 5:1; they were then stored in ethanol 70° at 4°C. Squashes of pollen mother cells were made using acetocarmine at 3%. Mitotic chromosome preparations were obtained from root-tips of germinating seeds. The material was pre-treated with 8-hydroxiquinoline 0.002 M for 4 h and fixed in ethanol:acetic acid (3:1); then for microscope observations it was stained following the Feulgen technique.
Nomenclature used for the karyotype description was that suggested by Levan et al. (Citation1964). The chromosome morphology was determined using the centromeric index (CI = short arm × 100/total chromosomal length). The chromosomes were classified as metacentric (m): 50–37.5 or submetacentric (sm): 35.5–25. The ideograms and chromosome measures were estimated from 10 metaphases plates of 7–10 individuals per species.
The karyotypes of V. loretensis (2n = 34), V. lucida (2n = 34), V. nudiflora (2n = 34), V. oligactoides (2n = 34), V. pseudolinearifolia (2n = 34), V. squamulosa (2n = 34), and V. tweediana (2n = 34) were carried out from a population analyzed by Vega and Dematteis (Citation2012).
The cluster analysis of karyotypic data was carried out in order to examine karyotype similarity among the species. A data matrix of eight OTUs (operational taxonomic units) and 10 variables was constructed. The variables were: total length of karyotype (TKL), the mean chromosome length (ML), size of the smallest (S) and size of the longest (L) chromosomes, the average CI and the ratio between the longest and shortest chromosome pair (R), and the number of m and sm chromosomes. The karyotype asymmetry has been determinate using intrachromosomal (A1) and interchromosomal (A2) index as suggested by Romero Zarco (Citation1986).
To calculate the average taxonomic distance Euclidia coefficent was used and to generate clustering method UPGMA was performed. Additionally, principal component analysis (PCA) was used to evaluate the contribution of each karyotypic parameter to the ordination of species.
Results
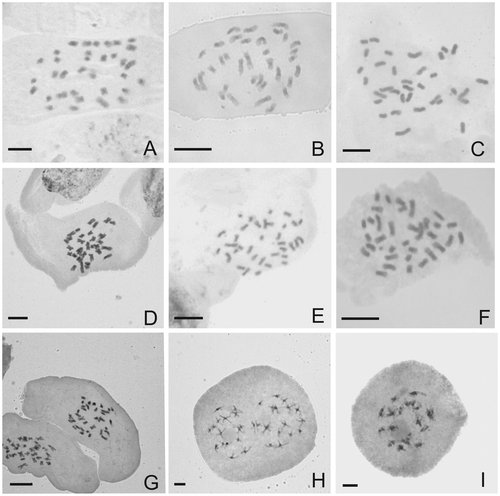
The number and behavior of meiotic chromosomes were determined for two populations of V. nudiflora (2n = 17II) and one population of V. chaquensis (2n = 17II) and V. tweediana (2n = 17II). The somatic chromosome numbers are reported for Vernonanthura brasiliana (2n = 34), V. chamaedrys (2n = 34), V. chaquensis (2n = 34) and Vernonia incana (2n = 34). The first chromosome count is presented for V. chaquensis (2n = 17II, 2n = 34). Figure shows photographs of the somatic and meiotic chromosomes.
Figure 1. (A–G) Somatic chromosomes of Vernonanthura and Vernonia incana; (H–I) meiotic chromosomes of Vernonanthura. (A) V. brasiliana, 2n = 2x = 34; (B) V. chaquensis, 2n = 2x = 34; (C) V. nudiflora, 2n = 2x = 34; (D) V. chamaedrys, 2n = 2x = 34; (E) V. squamulosa, 2n = 34; (F) V. incana, 2n = 34; (G) V. pseudolinearifolia, 2n = 34; (H) P II of V. chaquensis with 17I-17I chromosome; (I) diakinesis of V. tweediana with 17 II.
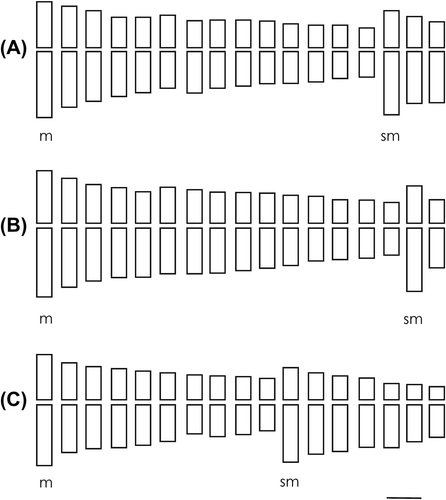
The karyotypes were determined for Vernonanthura chamaedrys (20m + 14sm), V. loretensis (26m + 8sm), V. lucida (32m + 2sm), V. nudiflora (30m + 4sm), V. oligactoides (24m + 10sm), V. pseudolinearifolia (28m + 6sm), V. squamulosa (30m + 4sm) and V. tweediana (20m + 14sm). For V. lucida (32m + 2sm), V. oligactoides (24m + 10sm) and V. pseudolinearifolia (28m + 6sm) they are presented for the first time. The ideograms of different species are shown in Figures and .
Figure 2. Idiograms of Vernonanthura species. (A) V. chamaedrys, 24m + 10sm; (B) V. loretensis, 26m + 8sm; (C) V. lucida, 32m + 2sm; (D) V. nudiflora, 30m + 4sm; (E) V. oligactoides, 24m + 10sm. Scale 1 μm.
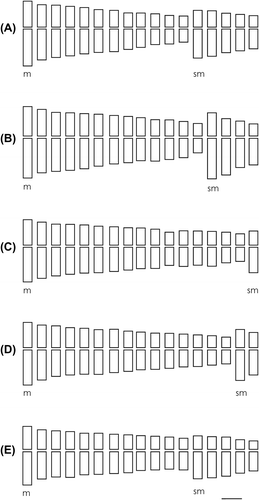
Figure 3. Idiograms of Vernonanthura species. (A) V. pesudolinearifolia, 28m + 6sm; (B) V. squamulosa, 30m + 4sm; (C) V. tweediana, 20m + 14sm. Scale 1 μm.
All stages of meiosis were stable without chromosomal irregularities in two population of V. nudiflora and V. chaquensis while V. tweediana presented 0.62% of lagging chromosomes in anaphase II, 0.30% of micronucleus in telophase I and 1.78% of bridges in anaphase I.
The karyotype formula, total length of karyotype, average chromosome length, range of chromosome length, centromeric index and asymmetry indexes of each species are given in Table . In all analyzed species, the karyotypes are composed of m and sm chromosomes.
Table 2. Karyotype formula, total karyotype length (TKL), mean chromosome length (ML), average centromeric index (CI), and asymmetry indexes (A1 and A2) of eight species of Vernonanthura.
Total length of karyotype ranged from 31.67 μm to 40.48 μm, while the average chromosome length varied from 1.86 μm to 2.38 μm and the centromeric index varied from 38.61 to 42.99. Vernonanthura oligactoides had smaller variation of chromosome length within karyotype (1.42 μm), while in V. squamulosa it was larger (2.15 μm).
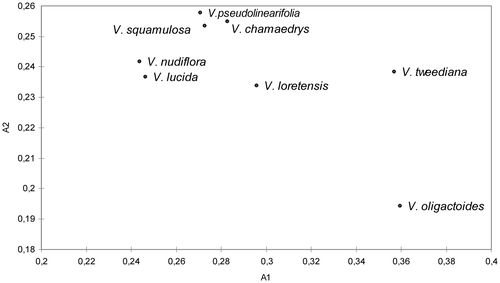
All taxa showed are relatively symmetrical in karyotype; A1 ranged from 0.2437 in V. nudiflora to 0.3593 in V. oligactoides and A2 varied from 0.1944 in V. oligactoides to 2.578 in V. pseudolinearifolia. The A1 and A2 indexes are plotted in a scatter diagram (Figure ).
Figure 4. Dispersion diagram representing the relations between the A1 and A2 karyotype asymmetry indexes.
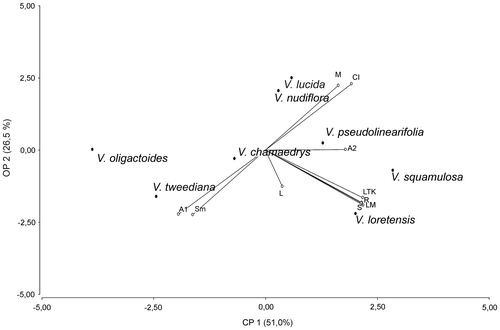
The UPGMA shows three clusters (Figure ). Cluster A, composed of V. nudiflora and V. lucida, was characterized by the highest CI and the lowest A1. Cluster B was formed by V. squamulosa, V. pseudolinearifolia and V. loretensis, the first two of which are grouped by ML, average CI and A1 and A2 indexes; within this cluster V. loretensis was separated by the greater phenetic distance. Cluster C was composed of V. chamaedrys, V. tweediana and V. oligactoides, which are characterized by a lower proportion of m chromosomes, higher proportion of sm chromosomes and a higher A1. The PCA of the karyotypic parameters shows that the first two principal components account for the 77% of the total variation; these are projected in a two-dimensional graphic (Figure ). Component one (61%) emphasizes R, S, ML and TKL, while component two (26%) accentuates variation in CI, proportions of m and sm chromosomes, and A1.
Figure 5. Dendrogram showing the phenetic relationships among the studied species of. Vernonanthura, constructed using the matrix of karyotype similarities with UPGMA. Cophenetic correlation coefficient r = 0.757. Abbreviations: Nud, V. nudiflora; Luc, V. lucida; Squa, V. squamulosa; Pseu, V. pseudolinearifolia; Lor, V. loretensis; Oli, V. oligactoides; Twee, V. tweediana; Cha, V. chamaedrys.
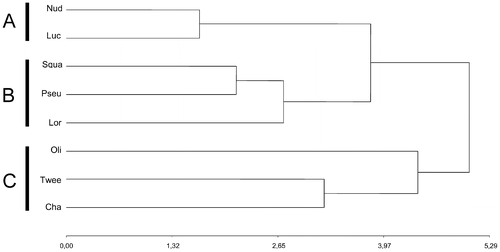
Figure 6. Diagram of principal component analysis of Vernonanthura species. Abbreviations: total length of karyotype (TKL), mean chromosome length (ML), size of the smallest (S) and size longest (L) chromosomes, average centromeric index (CI) and ratio between the longest and shortest chromosome pair (R), number of metacentric (m) and submetacentric chromosome (sm).
Discussion
All the analyzed species were diploid with basic number x = 17, which agrees with previous records (Hunter Citation1964; Jones Citation1974, 1979, Citation1982; Galiano and Hunziker Citation1987; Stutts Citation1988; Dematteis Citation1996, Citation1997, Citation2002; Dematteis and Fernández Citation1998; Carr et al. Citation1999; Watanabe et al. Citation2007; Salles de Melo et al. Citation2010; Vega and Dematteis Citation2012). The genus Vernonanthura has the basic number x = 17 and almost all its species are diploid with 2n = 34, excepting V. pinguis (Griseb.) H. Rob., which is tetraploid with 2n = 68 (Dematteis and Fernández Citation1998; Dematteis Citation2002).
Jones (Citation1979) proposed that all the New World Vernonieae have the basic chromosome number x = 17. However more recent studies (Ruas et al. Citation1991; Dematteis and Fernández Citation1998, Citation2000; Angulo and Dematteis Citation2012), indicate that x = 16 is the more frequent basic number in South America. Most species with x = 16 belong to Lessingianthus H. Rob., one of largest genera of Vernonieae in the New World, with more than 130 taxa widely distributed in southeastern Brazil, Paraguay, Uruguay, Bolivia and Argentina (Robinson Citation1999). It is cytologically characterized by the basic number x = 16 and possesses the greatest proportion of polyploid entities (Dematteis Citation2002).
Chromosome numbers are useful to distinguish the different South American genera of the tribe Vernonieae. Species belonging to Chrysolaena presents basic number x = 10, a base commonly found in Old World members of the tribe (Dematteis Citation2009). Lessingianthus presents a basic number x = 16, while Vernonanthura present the basic number x = 17. Lepidaploa is the only heterogeneous group showing four basic numbers, x = 14, 15, 16 and 17 (Dematteis Citation2002).
Vernonanthura is a cytologically homogeneous genus; almost all species are diploid with 2n = 34, excepting V. pinguis which is the only tetraploid taxon with 2n = 68 (Dematteis and Fernández Citation1998; Dematteis Citation2002). Vernonia sen. str. has only two South American species, V. incana is diploid with 2n = 34 (Dematteis and Fernández Citation1998; Dematteis Citation2002), while V. echioides has two cytotypes, one diploid with 2n = 34 and the other tetraploid with 2n = 68 (Vega and Dematteis Citation2012). The low variation in ploidy levels seems to be characteristic of these two genera because other South American members of Vernonieae are highly variable. In Chrysolaena there are species such as C. platensis (Spreng.) H. Rob. with cytotypes 2x, 4x, 6x, 8x; C. flexuosa (Sims) H. Rob. has 2x, 4x while that C. cognata (Less.) Dematt. has 2x, 4x, 5x, 6x and 8x (Dematteis Citation2002; Via do Pico and Dematteis Citation2012). In Lessingianthus it has been observed that L. rubricaulis and L. sellowii have 2x and 4x cytotypes (Angulo and Dematteis Citation2012).
Another type of numerical variation of chromosomes is the occurrence of accessory or B chromosomes, which are relatively frequent in the Asteraceae family (Jones Citation1995), including in species of Chrysolaena (Via do Pico and Dematteis Citation2012), Lessingianthus (Angulo and Dematteis Citation2009a, 2012), Mikania (Ruas et al. Citation2000), Crepis (Jamilena et al. Citation1994), and Haplopappus (Jackson and Newmark Citation1960). So far, the presence of these accessory chromosomes has not been documented in Vernonanthura and Vernonia (Stutts Citation1988; Dematteis and Fernández Citation1998; Dematteis Citation2002).
In Asteraceae dysploidy occurs in 214 genera, 21.9% of the 978 genera with counts reported (Semple and Watanabe Citation2009) and probably played an important role in the speciation mechanism of Vernonieae (Stebbins Citation1971; Dematteis Citation1996, 1998, 2002; Mansanares et al. Citation2002). This process was shown by Oliveira et al. (Citation2007) in the genus Vernonanthura, where V. polyanthes present populations 2n = 32, 2n = 34 and 2n = 36 in specimens from Brazil (Coleman Citation1968; Ruas et al. Citation1991; Oliveira et al. Citation2007).
According to Guerra (Citation1988) dysploidy and polyploidy are types of numerical chromosome variations that reflect phylogeny and karyotype evolution in plants. On the other hand variations such as aneuploidy and the presence of B chromosomes have clear implications beyond the species level. In Vernonia and Vernonanthura there are few records of these phenomena.
One of the most studied species of the genus has been Vernonanthura nudiflora and almost all the reports indicate 2n = 34 (Jones Citation1974; Bernardello Citation1986; Stutts Citation1988; Ruas et al. Citation1991; Dematteis and Fernández Citation1998; Angulo and Dematteis Citation2009a; Vega and Dematteis Citation2012). However, these results disagree with a prior analysis carried out by Covas and Hunziker (Citation1954) on a population of this species from western Argentina that recorded n = 16II (2n = 32). Although this chromosome number report could be attributed to an incorrect species identification, it is most likely due to the small size of species chromosomes, which in some cases contributes to the disparity of reported chromosome numbers (Guerra Citation1988) or to the occurrence of dysploidy in V. polyanthes (Oliveira et al. Citation2007).
Chromosomes of Vernonanthura species in general show a small size according to the classification suggested by Lima de Faria (Citation1980), with an average of 2.05 μm, which is consistent with previous studies (Ruas et al. Citation1991; Dematteis and Fernández Citation1998; Oliveira et al. Citation2007). The chromosome size is a feature that distinguishes genera and species of South American Vernonieae. Chrysolaena has an average size of chromosomes that varies between 2.4 and 2.5 μm (Dematteis Citation1997, 2009), while Lessingianthus, a related genus, has an average size of 1.75 μm (Angulo and Dematteis Citation2009a, 2009b). It can thus be assumed that during the evolution of the South American Vernonieae changes may have occurred in the chromosome size. These variations may be due to reciprocal translocations, thereby resulting in a decrease in the size of the chromosomes (Metel et al. Citation2004). The karyological information available indicates that the most primitive species (x = 10) have comparatively larger chromosomes than more advanced taxa (Ruas et al. Citation1991; Dematteis Citation1997; Dematteis and Fernández Citation1998). Species with base number x = 17 have an average size of 1.9–2.0 μm (Dematteis and Fernández,Citation1998), while species with x = 14 and x = 15 show an average length of 1.3–1.4 μm (Dematteis Citation1996; Dematteis and Fernández Citation1998).
All the analyzed species showed similar chromosomal features, although some differences in karyotype formula, chromosome size and karyotype asymmetry can be observed. The interchromosomal and intrachromosomal asymmetry index showed some differences among all species, except Vernonanthura chamaedrys, V. pesudolinearifolia and V. squamulosa. These species have similar asymmetry index, but they present different karyotype formulae (V. chamaedrys, 24m + 10sm; V. pseudolinearifolia, 28m + 6sm and V. squamulosa, 30m + 4sm). Species with lower values of A1 (V. lucida; V. nudiflora; V. pseudolinearifolia and V. squamulosa) tend to have a greater number of metacentric chromosomes. In regard to A2, all species have low values, suggesting that there is little variation in the size of the chromosomes within each species.
Vernonanthura chamaedrys and V. tweediana have the same karyotype formula (20m + 24sm) and similar total karyotype length (V. chamaedrys, 33.83 μm and V. tweediana, 33.23 μm) and average chromosome length (V. chamaedrys, 1.99 μm and V. tweediana, 1.95 μm). However, they are different in chromosome size (V. chamaedrys, 1.91 μm and V. tweediana, 1.75 μm), centromeric index (V. chamaedrys, 41.64 μm and V. tweediana, 38.61 μm) and intrachromosomal index (V. chamaedrys, 0.2826 and V. tweediana, 0.3568). Both species are easily recognized in the field because V. tweediana is a shrub around 220 cm tall with lanceolate leaves, while V. chamaedrys is a subshrub around 80–150 cm with ovate-oblong leaves.
The same karyotype formula was observed in Vernonanthura nudiflora and V. squamulosa (30m + 4sm), but they differ in other karyotype characteristics: V. squamulosa is longer in the total length of karyotype (37.99 μm) than V. nudiflora (33.53 μm), and consequently V. squamulosa has the longest average chromosome length. Additionally, they are easily recognized by their morphology and geographical distribution. Vernonanthura nudiflora has linear leaves, pedunculate heads and involucre 7–10 mm long, while V. squamulosa has ovate-lanceolate leaves, shortly pedunculate heads and involucre 9–10 mm long (Cristóbal and Dematteis Citation2003).
The karyotypes of V. lucida (32m + 2sm), V. oligactoides (24m + 10sm) and V. pseudolinearifolia (28m + 6sm) are presented for first time. The chromosome number of V. pseudolinearifolia was reported in the first cytological study of this species (Vega and Dematteis Citation2012).
The chromosome complement of V. chamaedrys was analyzed by Dematteis (Citation1996) and Dematteis and Fernández (Citation1998). In both cases, the karyotype formula was 22m +12sm, which does not coincide with the results obtained here, where this species presented a karyotype formula of 24m + 10sm. A previous analysis in V. loretensis showed a karyotype formula of 20m + 14sm (Dematteis Citation1996); meanwhile the results obtained in this work show that this species presents 26m + 8sm. Vernonanthura nudiflora is one of the most cytologically studied species; and different karyotype formula have been observed: 28m + 6sm (Ruas et al. Citation1991; Angulo and Dematteis Citation2009a) and 24m + 10sm (Dematteis Citation1997; Dematteis and Fernández Citation1998).The results obtained here (30m + 4sm) show a slight difference to those obtained by Ruas et al. (Citation1991) and Angulo and Dematteis (Citation2009b). The karyotype formula of V. squamulosa (30m + 4sm) does not match the previous analysis by Dematteis and Fernández (Citation1998), who proposed a karyotype formula of 20m + 12sm + 2 m-sm.
The UPGMA analysis demonstrated certain congruence between karyotype analysis and classification based on morphological characters. Cluster A contains V. nudiflora and V. lucida, which have been traditionally grouped under Vernonia section Lepidaploa subsect. Nudiflorae (Cabrera Citation1944). On the other hand, cluster B is composed of quite different species and the group does not reflect any previous classification. In cluster C, V. oligactoides and V. chamaedrys are morphologically similar and both were included under Vernonia section Lepidaploa subsection Chamaedrys by Cabrera and Klein (Citation1980) and Stutts (Citation1988).
PCA allowed observation of the main differences between Vernonanthura species with respect to different karyotype parameters. Three groups were recognized: V. oligactoides, V. chamaedrys, and V. tweediana were grouped in the lower left quadrant, characterized by a high values of A1 and a high proportion of sm chromosomes; V. loretensis and V. squamulosa were grouped in the lower right quadrant with high R, ML, S and TKL; and V. lucida, V. nudiflora and V. pseudolinearifolia, with high values CI and a high proportion of m chromosomes, were located toward the positive side of APC1 and ACP2. In this analysis, V. pseudolinearifolia is closer to this group and not to group B as in UPGMA analysis.
The results obtained in this work show that there are differences between the karyotypes of Vernonanthura species that may be utilized in taxonomic and evolutionary studies.
References
- Angulo MB, Dematteis M. 2009a. Karyotype analysis in eight species of Vernonia (Vernonieae, Asteraceae) from South American. Caryologia. 62(2):81–88.
- Angulo MB, Dematteis M. 2009b. Caryological analysis of South American species of Vernonia (Vernonieae, Asteraceae). Pl Biosyst. 142(1):20–24.
- Angulo MB, Dematteis M. 2012. Cytotaxonomy of some species of the South American genus Lessingianthus (Asteraceae, Vernonieae). Plant Syst Evol. 298(2):277–285.
- Bernardello LM. 1986. Números cromosómicos en Asteraceae de Córdoba. Darwiniana. 6(3):265–379.
- Bremer K. 1994. Asteraceae. Cladistics and classification. Portland (OR): Timber Press.
- Cabrera AL. 1944. Vernonias Argentinas (Compositae). Darwiniana. 6(3):265–379.
- Cabrera AL, Klein RM. 1980. Compostas, Tribo Vernonieae. In: Reitz R, editor. Flora Catarinense. Itajaí: Santa Catarina; p. 227–408.
- Carr GD, King RM, Powell AM, Robinson H. 1999. Chromosome numbers in Compositae XVIII. Amer J Bot. 86(7):1003–1013.
- Coleman JR. 1968. Chromosome numbers in Brazilian Compositae. Rhodora. 70(782):228–379.
- Covas G, Hunziker J. 1954. Estudios cariológicos en Antófitas IV parte. Revista Invest Agríc. 8(3):249–253.
- Cristóbal CL, Dematteis M. 2003. Asteraceae, Tribu I. Vernonieae. In: Ariza Espinar L, editor. Fl. Fanerog. Argentina. Cordoba: Museo Botánico. pp. 3–53.
- Dematteis M. 1996. Estudios cromosomicos en especies argentinas de Vernonia (Asteraceae). Bonplandia. 9(1–2):103–110.
- Dematteis M. 1997. Cromosomas en Vernonia platensis y especies afines (Asteraceae). Bonplandia. 9(3–4):259–264.
- Dematteis M. 1998. Karyotype analysis in some Vernonia species (Asteraceae) from South America. Caryologia. 51(3–4):279–288.
- Dematteis M. 2002. Cytotaxonomic analysis of South American species of Vernonia (Vernonieae: Asteraceae). Bot J Linn Soc. 139(4):401–408.
- Dematteis M. 2009. Revisión taxonómica del género sudamericano Chrysolaena (Vernonieae, Asteraceae). Bol Soc Argent Bot. 44(1–2):103–170.
- Dematteis M, Fernández A. 1998. Karyotypes of seven South American species of Vernonia (Asteraceae). Cytologia. 63 (3):323–328.
- Dematteis M, Fernández A. 2000. Chromosome studies on nine South American species of Vernonia (Vernonieae, Asteraceae). Caryologia. 53(1):55–61.
- Galiano NG, Hunziker JH. 1987. Estudios cariológicos en Compositae. IV. Vernonieae y Eupatorieae. Darwiniana. 28(1–4):1–8.
- Guerra M. 1988. Introdução a Citogenética Geral. 6th ed. Rio de Janeiro: Guanabara Koogan; p. 190–193.
- Hunter GE. 1964. Chromosome numbers in Vernonia: Section Lepidaploa Subsection Paniculatae verae. Southwest Nat. 9(4):239–244.
- Jackson RC, Newmark KP. 1960. Effects of supernumerary chromosomes on production of pigment in Haplopappus gracilis. Science. 132(3436):1316–1317.
- Jamilena M, Riuz Rejon C, Ruiz Rejon M. 1994. A molecular analysis of the origin of the Crepis capillaris B chromosome. J Cell Sci. 107(3):703–708.
- Jones SB. 1974. Vernonieae (Compositae) chromosomes numbers. J Torrey Bot Soc. 101(1):31–34.
- Jones SB. 1979. Chromosome numbers of Vernonieae (Compositae). B Torrey Bot Club. 106(2):79–84.
- Jones SB. 1982. In: Löve A, ed. IOPB chromosome number reports LXXIV. Taxon 31:126–127.
- Jones NR. 1995. B chromosomes in plants. Review. New Phytol. 131(85):411–434.
- Keeley SC, Robinson H. 2009. Vernonieae. In: Funk VA, Sunanna A, Stuessy T, Bayer RJ, editors. Systematic, evolution and biography of Compositae. Vienna: International Association for Plant Taxonomists; p. 439–469.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52(2):201–220.
- Lima de Faria A. 1980. Classification of genes, rearrangements and chromosomes according to the field. Hereditas. 93(1):1–46.
- Mansanares ME, Forni-Martins ER, Semir J. 2002. Chromosome numbers in the genus Lychnophora Mart. (Lychnophorinae, Vernonieae, Asteraceae). Caryologia. 55(4):367–374.
- Metel EV, Pocet V, Lamy F, Siljak-Yalolev S, Leujene B, Sar A. 2004. Chromosome evolution of Pennisetum species (Poaceae): implications of ITS phylogeny. Plant Syst Evol. 249(3):139–149.
- Oliveira MV, Forni- Martins ER, Semir J. 2007. Cytotaxonomic studies in six species of Vernonia (Asteraceae, Vernonieae). Caryologia. 60(1–2):7–47.
- Robinson H. 1992. A new genus Vernonanthura (Vernonieae: Asteraceae). Phytologia. 73(2):65–76.
- Robinson H. 1995. New combinations and new species in American Vernonieae (Asteraceae). Phytologia. 78(5):384–399.
- Robinson H. 1999. Generic and subtribal classification of American Vernonieae. Smithsonian Contr Bot. 89(1):1–116.
- Romero Zarco C. 1986. A new method for estimating karyotype asymmetry. Taxón. 35(3):526–530.
- Ruas PM, Ruas CF, Maffei EMD, Marin Morales MAM, Aguiar Perecin LR. 2000. Chromosome studies in the genus Mikania (Asteraceae). Gen Mol Biol. 23(4):979–984.
- Ruas PM, Ruas CF, Vieira AOS, Matzenbacher NI, Martins NS. 1991. Cytogenetics of genus Vernonia Schreber (Compositae). Cytologia. 56(2):239–247.
- Salles de Melo MRC, de Luca RM, Semir J, de Carvalho R, Araujo Pereira RC, Benko-Iseppon AM. 2010. Karyological features and cytotaxonomy of the tribe Vernonieae (Asteraceae). Plant Syst Evol. 285(3–4):189–199.
- Semple JC, Watanabe K. 2009. A review of chromosome numbers in Asteraceae with hypotheses on chromosomal base number evolution. In: Funk VA, Sunanna A, Stuessy T, Bayer RJ, editors. Systematic, evolution and biography of Compositae. Vienna: International Association for Plant Taxonomists; p. 439–469.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold.
- Stutts JG. 1988. Taxonomic revision of Vernonia Subsect. Chamaedrys (Compositae: Vernonieae). Rhodora. 90(861):37–99.
- Vega AJ, Dematteis M. 2008. Eupatorium rufescens y Vernonia oligactoides (Asteraceae), nuevas citas para la flora Argentina. Bonplandia. 17(1):83–89.
- Vega AJ, Dematteis M. 2011a. Nuevas combinaciones y tipificaciones en el género Vernonanthura (Vernonieae, Asteraceaea). Bol Soc Argent Bot. 46(3–4):369–374.
- Vega AJ, Dematteis M. 2011b. Pollen morphology of some species of Vernonathura (Asteraceae, Vernonieae) from southern South America. Palynology. 35(1):94–102.
- Vega AJ, Dematteis M. 2012. Chromosome studies of some species of Vernonanthura and Vernonia (Asteraceae, Vernonieae). Caryología. 65(4):271–275.
- Via do Pico GM, Dematteis M. 2012. Karyotype analysis and DNA content in some species of Chrysolaena (Asteraceae, Vernonieae). Pl Biosys. 147(4):864–863.
- Watanabe K, Yahara T, Hashimoto G, Nagatani Y, Soejima A, Kawahara T, Nakazawa M. 2007. Chromosome numbers and karyotypes in Asteraceae. Ann Missouri Bot Gard. 94(1):643–654.
