Abstract
Chromosome number, morphology and location of the nucleolus organizer region (NOR) are useful cytological characters for taxonomic and evolutionary studies. The karyotype of Upeneus moluccensis from the north-eastern Mediterranean was investigated through conventional Giemsa staining, Ag-NOR staining, C-banding and GTG-banding techniques. The results showed 2n = 44 (FN = 46), 2m + 2st + 40a chromosomes and four NORs. Two telomeric C-band regions were observed on a metacentric and subtelocentric chromosome pair. On the basis of the chromosome morphology, the GTG and C-banding pattern, it is hypothesized that Upeneus moluccensis is more primitive than the Mediterranean native fish species, Mullus barbatus and Mullus surmuletus. In this study, the cytogenetic characterization of U. moluccensis has been carried out for the first time.
Introduction
Understanding ecosystem biodiversity is fundamental to ecological research and key to maintaining a healthy environment, and for sustainable use of ecosystem services (Hajibabaei et al. Citation2011). Genetic variation is a fundamental characteristic of most biota and karyotype data have shown extensive variability between different species and higher taxonomic categories, proving a useful tool to indicate the evolutionary distance between species (Dobigny et al. Citation2004; Cioffi et al. Citation2012). Fish exhibit the greatest biodiversity among the vertebrates, making this group extremely appealing for the investigation of many evolutionary questions. Chromosomal studies with fishes from different regions of the world have provided reliable information on the inherent diversity of this group. Cytogenetics is a powerful tool for discovering biodiversity, with useful applications in evolutionary, taxonomic, phylogenetic and conservation studies (Cioffi et al. Citation2012).
Classical conventional karyotyping methods have been used for many years to determine chromosome number and morphology. The advent of chromosome banding techniques [i.e., centromere (C-), giemsa (GTC-) and silver staining-nucleolar organiser regions (AgNORs)] enabled the differentiation of specific regions along chromosomes. Nucleolar organizer regions (NORs) are particularly significant in chromosomal evolutionary analyses (Fujiwara et al. Citation1998; Affonso et al. Citation2002; Sato and Nishida Citation2010; Sczepanski et al. Citation2010). Highly polymorphic numbers and positions of NORs have been described in fish species (Phillips et al. Citation1988; Castro et al. Citation1996). The most common NOR localizations in fishes are reported at telomeric regions (Gold Citation1984; Karahan and Ergene Citation2010), with some taxa bearing NORs on a single chromosome pair (Galetti et al. Citation1984; Accioly and Molina Citation2008) and others showing a multi-chromosomal NOR distribution (Foresti et al. Citation1981; Phillips et al. Citation1988; Castro et al. Citation1996; Karahan and Ergene Citation2011).
In most eukaryotes, the centromere’s DNA sequence consists of large arrays of repetitive DNA (Mehta et al. Citation2010). C-banding stains the constitutive heterochromatin region, corresponding to repetitive regions in the genome, which are extremely condensed during the metaphase phase of cell division. Heterochromatin has played an important role in the study of the diversification of some fish groups (Molina et al. Citation1998; Margarido and Galetti et al. Citation2000; Hughes and Hawley Citation2009). It is associated with rearrangements, quantitative variation and the formation of new karyotypes (Miklos and Gill Citation1982; Rocco et al. Citation2002).
The Mullidae family (goatfishes) consists of 87 economically important species belonging to six genera (Eschmeyer and Fricke Citation2015); they inhabit coastal waters, always near the bottom (Froese and Pauly Citation2015). One of the largest genera of the family Mullidae is Upeneus Cuvier 1829, which comprises 37 species (Froese and Pauly Citation2015). Whereas most members of the genus originate from the Indian Ocean, there are a few species which derive from the Pacific and Atlantic oceans. Seven species of the Mullidae family are reported for the Mediterranean Sea, three of them native [Mullus barbatus Linnaeus, 1758; Mullus surmuletus Linnaeus, 1758 and Pseudupeneus prayensis (Cuvier, 1829)], three introduced [Upeneus pori Ben-Tuvia and Golani, 1989; Upeneus moluccensis (Bleeker, 1855) and Upeneus asymmetricus Lachner, 1954] and one of questionable origin [Parupeneus forsskali (Fourmanoir and Gueze, 1976)] (Froese and Pauly 2015). Upeneus moluccensis (goldband goatfish) is one of the most commercially viable (Nelson Citation2006) and successful Lessepsian fish species in the Levantine Basin.
In the present study, the karyotype of U. moluccensis was analysed using conventional Giemsa staining, C-banding, GTG-banding and AgNOR staining techniques. In traditional taxonomy, morphology is a key factor for describing and naming species, based on the physical characteristics such as size, shape, colour and anatomical structure. Karyological data could be useful in providing new elements for discussion in addition to those of traditional morphology. The present study was undertaken to provide the first analysis, to our knowledge, of the karyotype of U. moluccensis.
Methods
Samples were collected between January and December 2010 from the north-eastern Mediterranean; Mersin Bay (36°33′N, 34°15′E) and Iskenderun Bay (36°43′N, 35°47′E), and the Yesilovacik region (36°10′N, 33°37′E), with the Research Vessels Bilim-2 and Lamas using a trawl net. 59 U. moluccensis individuals (24 from Mersin Bay, 17 from Iskenderun Bay and 18 from Yesilovacik region) were used for cytogenetic analyses.
Leukocyte cell culture was applied to obtain metaphase chromosomes from the species blood cells. Mitotic chromosome preparation from lymphocyte cultures followed the protocol described by Fujiwara et al. (Citation2001). Caudal vein areas of individuals were cleaned with distilled-water soaked cotton and after complete drying swabbed with absolute ethanol. Approximately 1–2 ml blood was withdrawn from the caudal vein through a 2 ml syringe equipped with a 25Gx 5/8-inch needle pre-flushed with sodium heparin (Nevparin, MN, Istanbul, 25.000IU/5 ml). The blood was immediately transferred to sterile centrifuge tubes and stored at 4°C until use. Cell cultures were established within 24 h after blood withdrawal. After centrifugation of whole blood (2500 rpm for 10 min), leukocyte cells were collected from the interface between red cells and plasma and incubated in 5 ml medium (Biochrom, AG, Berlin) containing antibiotic and antimycotic (Sigma-Aldrich, Germany, 10,000 units penicillin, 10 mg streptomycin, and 25 μg amphotericin B per ml) for 72 h at 27°C. Following the incubation period colcemid (Biological Industries, Kibbutz Beit Haemek, Israel, 10 μg ml–1) was added to the cultures 3 h before harvesting. Despite the high amount of the antibiotic and antimycotic solutions (10–20 ml l–1), fungus contaminations occurred in 49-cell culture tubes. For this reason, metaphase plates could only be obtained from a total of 10 females of Mersin (six) and Iskenderun (two) bays and Yesilovacık (two) region.
Detection of the NORs was carried out following the silver (AgNO3) staining method of Howell and Black (Citation1980). Characterization of the pattern of constitutive heterochromatin was performed through the C-banding method (Sumner Citation1972). Chromosome morphology was determined according to the centromere position following the nomenclature by Levan et al. (Citation1964). Metaphase chromosomes were banded using the conventional trypsin-Giemsa banding (GTG-banding) technique (Seabright Citation1971). After ageing at 65°C for 45 min the metaphase chromosome slides were treated with a trypsin solution (0.25% in 1× PBS) for between 30 and 120 seconds. Trypsin-treated slides were rinsed in calf serum buffer (containing 500 μl FCS and 50 ml 0.025 M KH2PO4 buffer, pH 6.8) for 3 min followed by Giemsa staining (made in 0.025 M KH2PO4 buffer; pH 6.8) to visualize chromosomes. Chromosome arm number or fundamental number (FN) was determined considering subtelocentric and acrocentric (st/a) chromosomes as uni-armed, and metacentric and submetacentric (m/sm) chromosomes as bi-armed. MICRO-MEASURE software (http://micromeasure.software.informer.com/3.3/) was used to discriminate chromosomes types. Metaphase spreads were examined with a Nikon Optiphot-2 microscope equipped with a Nikon Digital camera (DXm1200F, Nikon Corp., Netherlands). Photographs were processed with the use of Adobe Photoshop software (v. CS5, http://www.adobe.com/tr/products/photoshop.html).
Results
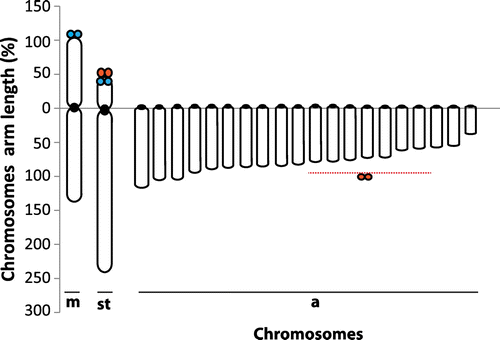
Of the 460 metaphase plates, 420 were counted as 2n = 44 with the karyotype of 2m + 2st + 40a chromosomes (FN = 46). The chromosome number of the other 40 metaphase plates was between 36 and 43. These metaphases were regarded as incomplete. Giemsa staining, C-banding, GTG-banding, AgNOR staining metaphase images and AgNOR staining interphase cell images are given in Figure a–e. Moderate heterochromatic blocks (C+ regions) were seen in the centromeric regions of 17 chromosome pairs, but conspicuous non-pericentromeric blocks (telomeric) were observed on the first metacentric chromosomes (Figures b, ). A subterminal heterochromatic segment was also observed on the short arm of chromosome pair 2 just above its terminal secondary constriction (Figures b, ). GTG-banding results showed certain positive bands for euchromatic and heterochromatic regions on the telomere regions of just three chromosome pairs, with one acrocentric pair almost entirely heterochromatic. On the other hand, no clear heterochromatic bands were seen for other chromosomes (Figure c).
Figure 1. Metaphase and interphase cell images of Upeneus moluccensis from the north-eastern Mediterranean: (a) Giemsa staining, (b) C-banding, (c) GTG-banding (d) AgNOR staining and (e) AgNOR interphase cell. Arrows indicate the first metacentric and second subtelocentric chromosomes with non-pericentromeric blocks, positive heterochromatic GTC regions and NORs respectively.
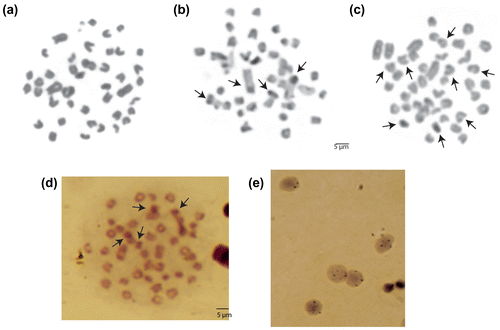
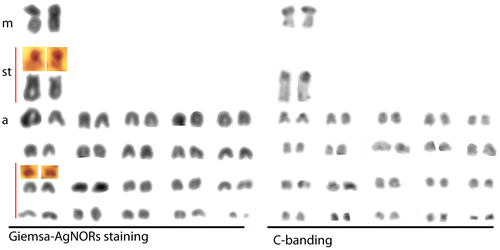
Figure 2. Giemsa staining, C-banding and AgNOR staining karyotypes of Upeneus moluccensis (2n = 44, 2m + 2st + 40a, FN = 46) from the north-eastern Mediterranean.
Four actively transcribed telomeric NORs regions were identified on the second subtelocentric chromosome pair and a small acrocentric chromosome pair (Figure d). During interphase, both ribosomal genes and their respective silver stained proteins are localized within the nucleolus (Hittmair et al. Citation1994). Carbajo et al. (Citation1993) reported that since both the mean area and numbers of AgNORs per cell increase from the G0–G1 phase to the early-mid and late-mid S-phases, the expression of AgNORs is causally or indirectly linked to DNA synthesis and, thus, AgNORs can be considered as a cell proliferation marker. In the present study interphase stage NORs numbers were observed between 2 and 4; with reference to the previous study we can say that “2” represents G0–G1, and “4” S or G2 phases (Figure e).
Karyotype images of the Giemsa staining, C-banding and AgNOR staining are presented in Figure . Arm ratios of chromosomes, non-centromeric C-band regions and NORs are given in Figure .
Discussion
In the present study, a cytogenetic analysis of U. moluccensis by conventional Giemsa staining, C-banding, GTG-banding and AgNOR staining has been carried out for the first time. Mullus barbatus and M. surmuletus are native fish species of the Mediterranean Sea and also the closest relatives of U. moluccensis in the area. The diploid chromosome numbers of both species were reported to be 44. The karyotype of M. barbatus was described as 6 m/sm + 16 st + 22t/a (FN = 50) with two NORs by Vitturi et al. (Citation1992) from Palermo (Sicily, Western Mediterranean), and by Saygun et al. (Citation2006) for the Zonguldak region (Western Black Sea). The karyotype of M. surmuletus was reported as 8m/sm + 16st + 20t/a (FN =52) and with two NORs by Vitturi et al. (Citation1992) from Palermo (Sicily, Western Mediterranean). In the present study, the chromosome number of U. moluccensis was also determined as 44 and the karyotype consisted of 2m + 2st + 40a chromosomes with four NORs and four telomeric positive heterochromatin regions (C-bands). Two NORs were reported at the terminal position on the short arm of a medium-sized a-/st chromosome for M. barbatus and M. surmuletus species by Vitturi et al. (Citation1992). In the present study four NORs were identified on the second subtelocentric and on a small acrocentric chromosome pair. The NORs on the acrocentric chromosomes are compatible with those of related species M. barbatus and M. surmuletus and were found in typical locations for fishes (Hartley Citation1987; Amemiya and Gold Citation1988).
Chromosome number and morphological variation among fish species can be used to investigate the evolutionary relationships among and within species (Thorgard and Disney Citation1990; Molina et al. Citation2008; Karahan and Ergene Citation2009, Citation2011). A chromosome number of 2n = 48, consisting entirely of uni-armed elements, has been suggested as primitive for teleostean fish (Ohno Citation1970, Citation1974; Chen Citation1971). When we focus on the karyotypes of M. barbatus, M. surmuletus (6 m/sm + 16 st + 22t/a and 8m/sm + 16st + 20t/a respectively) and the present study species (2m + 2st + 40a), given the knowledge that “the more uni-armed elements the more primitive” (Ohno Citation1970, Citation1974; Chen Citation1971; Greenbaum et al. Citation1978) we can say that U. moluccensis is more primitive than those native species. Additionally, the primitive karyotype was proposed to have little autosomal heterochromatin and this heterochromatin is probably limited to the centromeric regions (Greenbaum et al. Citation1978; Koop et al. Citation1984). In the present study, besides constitutive centromeric heterochromatin on 17 chromosome pairs and telomeric blocks on chromosome 1 only a few GTC-band regions were seen. Non-pericentromeric C-bands of U. moluccensis possibly indicate that these C-band sequences evolved during the diversification of the species, perhaps occupying distinct chromosome territories and being subject to chromosomal dispersion patterns (Dos Santos Abel et al. Citation2006) and karyotypic rearrangement (Galetti et al. Citation2000).
Despite their high economic value, mullid cytogenetics has received little attention to date, because of the both the intrinsic difficulties in analysing karyotypes with small chromosomes sizes and the problem of keeping fish alive after sampling. It is believed that this study will significantly contribute to mullid taxonomy research.
Funding information
This work was financed by the TUBITAK Small Pelagic Fishes project [1080566] and the TUBITAK National Post-Doctoral Research Scholarship Program.
Disclosure statement
No potential conflict of interest was reported by the author.
Acknowledgements
I would like to thank Associate Prof Dr Ali Cemal Gucu, Dr Yesim Ak Orek, Dr Meltem Ok and Dr Serdar Sakinan for their help during sampling and Ms Alison Kideys for correcting the manuscript.
References
- Accioly IV, Molina WF. 2008. Cytogenetic studies in Brazilian marine Sciaenidae and Sparidae fishes (Perciformes). Genet Mol Res. 7(2):358–370.
- Affonso PRAD, Guedes W, Pauls E, Galetti PM Jr. 2002. Close karyotypical relationship between two species of marine angelfishes from South Atlantic: Pomacanthus arcuatus and P. paru (Perciformes, Pomacanthidae). Caryologia. 55(4):323–329.
- Amemiya CT, Gold JR. 1988. Chromosomal NORs as taxonomic and systematic characters in North American cyprinid fishes. Genetica. 76(2):81–90.
- Carbajo S, Orfao A, Vicente-Villardon JL, Carbajo-Perez E. 1993. Expression of silver-stained nucleolar organizer regions is coupled to cell cycle in rat thymic cells. Cytometry. 14(1):46–52.
- Castro J, Viñas A, Sánchez L, Martínez P. 1996. Characterization of an atypical NOR site polymorphism in brown trout (Salmo trutta) with Ag- and CMA3-staining, and fluorescent in situ hybridization. Cytogenet Cell Genet. 75(4):234–239.
- Chen TR. 1971. A comparative chromosome study of twenty killifish species of the genus Fundulus (Teleostei:Cyprinodontidae). Chromosoma. 32(4):436–453.
- Cioffi MB, Molina WF, Artoni RF, Bertollo LAC. 2012. Chromosomes as Tools for Discovering Biodiversity – The Case of Erythrinidae Fish Family. In: Tirunilai Padma, editor. Recent Trends in Cytogenetic Studies - Methodologies and Applications. InTech. p. 125–146. Available from: http://cdn.intechopen.com/pdfs-wm/30743.pdf
- Dobigny G, Ducroz JF, Robinson TJ, Volobouev V. 2004. Cytogenetics and cladistics. Syst Biol. 53(3):470–484.
- Dos Santos Abel LD, Mantovani M, Moreira-Filho O. 2006. Chromosomal distribution of the As51 satellite DNA in two species complexes of the genus Astyanax (Pisces, Characidae). Genet Mol Biol. 29(3):448–452.
- Eschmeyer WN, Fricke R. 2015. Catalog of fishes: genera, species, references. Eschmeyer B, editor; [cited 2015 Sep 2]. Available from: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp
- Foresti F, Almeida Toledo LF, Toledo SA. 1981. Polymorphic nature of nucleolus organizer regions in fishes. Cytogenet Cell Genet. 31(3):137–144.
- Froese R, Pauly D. 2015. FishBase [Internet]; [cited 2015 Aug]. Available from: www.fishbase.org
- Fujiwara A, Abe S, Yamaha E, Yamazaki F, Yoshida MC. 1998. Chromosomal localization and heterochromatin association of ribosomal RNA gene loci and silver-stained nucleolar organizer regions in salmonid fishes. Chromosome Res. 6(6):463–471.
- Fujiwara A, Nishida-Umehara C, Sakamoto T, Okamoto N, Nakayama I, Abe S. 2001. Improved fish lymphocyte culture for chromosome preparation. Genetica. 111(1–3):77–89.
- Galetti PM Jr, Foresti F, Bertollo LAC, Moreira-Filho O. 1984. Characterization of eight species of Anostomidae (Cypriniformes) fish on the basis of the nucleolar organizing region. Caryologia. 37(4):401–406.
- Galetti PM Jr, Aguilar CT, Molina WF. 2000. An overview of marine fish cytogenetics. Hydrobiologia. 420(1–3):55–62.
- Greenbaum IF, Baker RJ, Ramsey PR. 1978. Chromosomal evolution and its implications concerning the mode of speciation in three species of deer mice of the genus Peromyscus. Evolution. 32(3):646–654.
- Gold JR. 1984. Silver-staining and heteromorphism of chromosomal nucleolus organizer regions in North American cyprinid fishes. Copeia. 1984(1):133–139.
- Hajibabaei M, Shokralla S, Zhou X, Singer GAC, Baird DJ. 2011. Environmental barcoding: A next-generation sequencing approach for biomonitoring applications using river benthos. PLoS ONE. 6(4):e17497.
- Hartley SE. 1987. The chromosomes of Salmonid fishes. Biol Rev. 62(3):197–214.
- Hittmair A, Offner F, Feichtinger H, Ensinger C, Rogatsch H, Mikuz G, Öfner D. 1994. In vitro investigations of interphase and metaphase argyrophilic nucleolar organizer regions and cellular proliferation in the human urothelial cancer cell line HOK-1. Virchows Archiv. 424(2):149–154.
- Howell WM, Black DA. 1980. Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 36(8):1014–1015.
- Hughes SE, Hawley RS. 2009. Heterochromatin: a rapidly evolving species barrier. PLoS Biol. 7(10):e1000233.
- Karahan A, Ergene S. 2009. Cytogenetic variation of geographically isolated four populations of Garra rufa [(Heckel, 1843) (Pisces, Cyprinidae)]. Caryologia. 62(4):276–287.
- Karahan A, Ergene S. 2010. Cytogenetic analysis of Garra variabilis (Heckel, 1843) (Pisces, Cyprinidae) from Savur Stream (Mardin). Turkey. Turk J Fish Aquat Sci. 10(4):483–489.
- Karahan A, Ergene S. 2011. Chromosomal differentiation between populations of Clarias gariepinus (Burchell, 1822) from the Göksu Delta and Orontes River (Turkey). Turk J Biol. 35(1):79–87.
- Koop BF, Baker RJ, Haiduk MW, Engstrom MD. 1984. Cladistical analysis of primitive G-band sequences for the karyotype of the ancestor of the Cricetidae complex of rodents. Genetica. 64(3):199–208.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52(2):201–220.
- Margarido VP, Galetti PM Jr. 2000. Amplification of a GC–rich heterochromatin in the freshwater fish Leporinus desmotes (Characiformes, Anostomidae). Genet Mol Biol. 23(3):569–573.
- Mehta GD, Agarwal MP, Ghosh SK. 2010. Centromere identity: a challenge to be faced. Mol Genet Genom. 284(2):75–94.
- Miklos GLG, Gill AC. 1982. Nucleotide sequences of highly repeated DNAs: compilation and comments. Genet Res. 39(1):1–30.
- Molina W, Schmid M, Galetti PM Jr. 1998. Heterochromatin and sex chromosomes in the neotropical fish genus Leporinus (Characiformes, Anostomidae). Cytobios. 94(377):141–149.
- Molina WF, Shibatta O, Galetti PM Jr. 2008. Chromosomal evidence of population subdivision in the freshwater fish Leporinus elongatus in the Upper Paraná River basin. Genet Mol Biol. 31(1):270–274.
- Nelson JS. 2006. Fishes of the world. Hoboken: John Wiley & Sons.
- Ohno S. 1970. The enormous diversity in genome sizes of fishes as a reflection of Nature's extensive experiments with gene population. Trans Am Fish Soc. 99(1):120–130.
- Ohno S. 1974. Protochordate, Cyclostoma and Pisces. In: John B, editor. Animal Cytogenetics. Vol. 4, Chordata 1 Berlin: Gebrüder Borntraeger. p.1–91.
- Phillips RB, Pleyte KA, Hartley SE. 1988. Stock-specific differences in the number and chromosome positions of the nucleolar regions in Arctic charr (Salvelinus alpinus). Cytogenet Cell Genet. 48(1):9–12.
- Rocco L, Morescalchi MA, Costagliola D, Stingo V. 2002. Karyotype and genome characterization in four cartilaginous fishes. Gene. 295(2):289–298.
- Sato Y, Nishida M. 2010. Teleost fish with specific genome duplication as unique models of vertebrate evolution. Environ Biol Fish. 88(2):169–188.
- Saygun S, Karayucel I, Bircan R. 2006. Karyological observation of red Mullet (Mullus barbatus Linnaeus, 1758). Turk J Biol. 30(4):235–238.
- Sczepanski TS, Noleto RB, Cestari MM, Artoni RF. 2010. A comparative study of two marine catfish (Siluriformes, Ariidae): cytogenetic tools for determining cytotaxonomy and karyotype evolution. Micron. 41(3):193–197.
- Seabright M. 1971. A rapid banding technique for human chromosomes. Lancet. 2(7731):971–972.
- Sumner AT. 1972. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 75(1):304–306.
- Thorgard GH, Disney JE. 1990. Chromosome preparation and analysis. Methods for fish biology. Bathesda, MD: American Fisheries Society.
- Vitturi R, Mazzola A, Catalano E, Lo Conte MR. 1992. Karyotype characterization of four Mediterranean sparid fish (Pisces, Perciformes) using conventional and banding techniques. Cytobaios. 72(289):107–115.