Abstract
Karyological characteristics were determined in 36 populations of eight Chara species and varieties distributed in Iran. The karyotype asymmetry/symmetry were evaluated using CVCL (coefficient of variation of chromosome length), CVCI (heterogeneity of the centromeric index) and MCA (mean centromeric asymmetry). Results obtained from the current study revealed that there are two different ploidy levels (n = 14 and n = 28) among the investigated taxa. All eight species and varieties had metacentric (M & m), sub-metacentric (Sm) and sub-telocentric (St) chromosomes, but the majority of taxa showed a dominance of metacentric and sub-metacentric chromosomes. In general, the karyotypes within the genus Chara were symmetrical, but the studied species differed significantly in quantitative features of karyotype and revealed the occurrence of qualitative changes during the species diversification. The present study reveals that karyological features may be of great help in Chara species delimitations.
Introduction
Charophytes, commonly known as stoneworts, muskgrasses or bassweeds, are a large group of macrophytic green algae that have a global distribution and mainly grow in submerged conditions of slow running and standing water over muddy and sandy substrates (Corillion Citation1975; Krause Citation1997; Casanova Citation2005). The organization of the body and life cycle of the Characeae are relatively complex (Casanova and Karol Citation2014). They are oogamous, have motile sperm cells, and possess sterile cells surrounding their antheridial filaments (forming a globule) as well as their oogonia (forming a nucule) (Bold and Wynne Citation1985). The plant body of charophytes is gametophyte and gametes (also n chromosomes) are generated via mitotic division in the reproductive structures. Doubling of the chromosomes occurs at fertilization. Thus, the zygote is believed to be the only diploid phase (sporophyte) in the life history (Wood and Imahori Citation1965). The occurrences of mitosis in the actively dividing antheridial cells make them the most favorable material for cytological study in charophytes.
The chromosome numbers reported for the charophyte genera revealed the significance of polyploidy (euploidy and aneuploidy), in the species diversification and evolution of this group. Many studies reported 14 as the basal haploid chromosome number for Chara, from which all other numbers were derived (Casanova Citation2015). Many of dioecious species have this chromosome number (Guerlesquin Citation1984), supporting an ancestral condition of n = 14. The number and type of the chromosomes (karyotype) can be an important clue to the separation of closely related taxa in Chara (Proctor Citation1971).
Karyological studies on Chara are often restricted to just reporting of chromosome numbers (Gillet Citation1959; Sarma and Khan Citation1965; Guerlesquin Citation1967b; Sarma and Ramjee Citation1971; Chatterjee Citation1976; Casanova Citation2015) whereas karyological analyses were rarely reported (Sawa Citation1965; Mukhopadhyay and Ray Citation1995; Khatun et al. Citation2009; Singh and Chaudhary Citation2010). The purposes of the present study were to report haploid chromosome numbers of several Chara populations from Iran to investigate their relationships based on karyotype features
Material and methods
Plant material
In order to study the karyological characteristics of Chara species, fertile plants with growing tips bearing young stages of gametangial development were collected from natural habitats, distributed in six provinces of Iran which differed in latitude, longitude and altitude (Table ). Five randomly collected specimens were investigated for each population. Karyological characteristics were determined in 36 populations of eight Chara species and varieties growing in Iran.
Table 1. Chara populations studied, their locality and voucher number.
The species and varieties studied are Chara vulgaris (var. longibracteata (Kützing) J. Groves & Bullock-Webster (four populations), Chara vulgaris var. vulgaris (Linnaeus) R. D. Wood (twelve populations), Chara gymnophylla (A. Braun) (six populations), Chara gymnophylla var. rohlenae (Vilhelm) Ahmadi in Ahmadi et al. Citation2012: 375, figs 33–38 (seven populations), Chara contraria A. Braun ex Kützing (two populations), Chara connivens Salzmann ex A. Braun (one population), Chara braunii Gmelin (two populations), and Chara globata Migula (one population).
At least five chromosome plates were analyzed per specimen. Voucher specimens have been deposited in the Herbarium of Shahid Beheshti University (HSBU).
Cytological studies
Antheridial filaments were separated for cytological study in charophytes. Green tips with prominent reproductive structures were collected and fixed in 1:3 acetic ethanol solution (Carnoy’s fluid) for 24–48 h and then transferred to 70% ethanol and stored in a refrigerator. Squashing technique was carried out for cytological studies with 2% aqueous aceto-orcein as the stain. Chromosomes number and karyotype details were studied in at least five well-prepared metaphase plates. The chromosome were photographed by using Olympus BX51 microscope with automatic camera and measured by Image Tools 3 software (Sheidai and Rashid Citation2007).
The chromosome counts are listed in Table . The numbers are gametic numbers taken from mitoses in the antheridial filaments prior to formation of the sperm. The chromosomes were identified according to the Levan method (Levan et al. Citation1964), and karyotype symmetry was determined according to Stebbins (Citation1971) and also by a new method described by Paszko (Citation2006) as follows: (1)
Table 2. Karyotype features of Chara species studied.
where SCI = the standard deviation of the centromeric index; xCI = the mean centromeric index, CVCL = A2 × 100. A2 is proposed by Romero Zarco (Citation1986) and Peruzzi and Eroğlu (Citation2013): (2)
where A is proposed by Watanabe et al. (Citation1999).
Karyotypic parameters such as the total form percentage (TF%) and coefficient of variation (CV) of the chromosome size were determined (Sheidai and Jalilian Citation2008).
A bidimensional scatter plot was drawn with parameters CVCL and MCA to reveal karyotype relationships among the studied species. An idiogram was constructed for each taxon.
Statistical analyses
The analysis of variance (ANOVA) and the least significant difference test (LSD) were performed to reveal significant differences in the total size of chromosomes, size of the long arms, size of the short arms, and the arm ratios among the populations of each species and among species with similar chromosome numbers (Sheidai and Rashid Citation2007).
Pearson coefficient of correlation was determined among the number of chromosomes, total size of chromosomes and the mean chromosome length in the studied species with their geographical features, viz. longitude, latitude and altitude (Sheidai et al. Citation1996).
Grouping of the species and populations with similar chromosome number was performed using various clustering methods including unweighted paired group with arithmetic mean (UPGMA) and the WARD (minimum spherical method), as well as ordination based on principal component analysis (PCA) (Podani Citation2000; Sheidai et al. Citation2002). PAST ver.3.01 (Hammer et al. Citation2013) was used for statistical analyses.
Results
The haploid chromosome number and karyotype features of the studied species are presented in Table . The haploid idiograms are given in Figure A–J.
Figure 1. Haploid idiograms and metaphase plate of Chara species and varieties: (A, B) C. vulgaris var. vulgaris (n =14 and n =28); (C, D) C. vulgaris var. longibracteata (n =14 and n =28); (E) C. gymnophylla var. rohlenae; (F) C. gymnophylla; (G) C. braunii; (H) C. contraria; (I) C. connivens; and (J) C. globate. Scale bars: 13 μm.
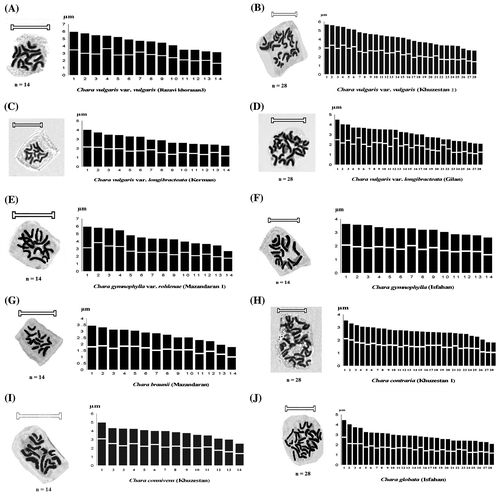
Twenty-five populations of eight species and varieties studied showed a chromosome number of n = 14 chromosome number, while the other showed n = 28.
The species studied differed in their karyotype formula (Table ). They had metacentric (M & m), sub-metacentric (Sm) and sub-telocentric (St) chromosomes. In all examined taxa, metacentric chromosomes were the most common form of chromosomes (87.3% of all chromosome) followed by submetacentric chromosomes (11.9%). The subtelocentric chromosomes were relatively few in number (0.79%).
The size of the largest chromosome varied from 6.52 μm in the Isfahan population of C. gymnophylla var. rohlenae to 2.48 μm in the Mazandaran population of C. gymnophylla. The size of the shortest chromosome varied from 3.55 μm in the Gilan population of C. gymnophylla to 1.16 μm in the Mazandaran population of C. gymnophylla.
The highest value of total haploid chromosome length occurred in the Khuzestan population of C. vulgaris var. vulgaris (113.34 μm), while the lowest value occurred in the Razavi Khorasan population of C. vulgaris var. vulgaris (27.60 μm).
The highest value of coefficient of variation (CV%) for the size of the chromosomes (28.41 μm) occurred in the Mazandaran population of C. vulgaris var. vulgaris, while the lowest CV value (9.42 μm) was observed in the Gilan population of C. gymnophylla. Total form percentage value (TF%) varied from 45.42 in the Mazandaran population of C. gymnophylla to 36.74 in the Kerman population of C. gymnophylla var. rohlenae. A higher value of TF% indicates the presence of relatively more symmetrical karyotype.
The Chara species were placed in 1A, 2A, 1B and 2B classes of Stebbins karyotype system; these are considered relatively primitive. Among the species that were placed in 2B class, the Isfahan population of C. gymnophylla var. rohlenae showed a higher value of Romero Zarco A1 index (0.38) and had relatively a more asymmetrical karyotype.
Asymmetry indices estimated on the basis of statistical data resolved the Chara karyotypes into the range of symmetrical to lowest asymmetrical values.
The Mazandaran population of C. vulgaris var. vulgaris was characterized by the highest value of CVCL and the Gilan population of C. vulgaris var. longibracteata was characterized by the highest value of CVCI. The Isfahan and Gilan populations of C. gymnophylla were characterized by lower values of both CVCL and CVCI respectively (Table ). According to the scatter plot (Figure ) obtained from the parameters CVCL versus MCA, C. gymnophylla and C. gymnophylla var. rohleana showed the lowest and highest MCA, respectively.
Figure 2. Scatter diagram of Chara species based on the karyotype parameters CVCL versus MCA. C. vulgaris var. vulgaris (), C. vulgaris var. longibracteata (■), C. gymnophylla var. rohleana (∆), C. gymnophylla (▲), C. braunii (*), C. contraria (×), C. connivens (+) and C. globata (■).
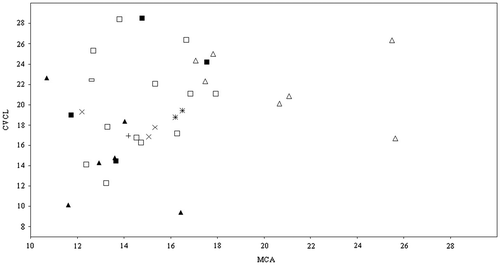
C. gymnophylla and C. vulgaris (var. vulgaris and var. longibracteata) demonstrated the lowest and the highest value for CVCL, respectively. As can be seen in Figure , the eight species and varieties of the Chara genus were separated based on asymmetry: for example C. braunii, C. contraria and C. connivens showed relatively low intrachromosomal (MCA) and interchromosomal asymmetry (CVCL).
Pearson correlation determined among the karyotype features and geographical features showed that the number of chromosomes, total size of chromosome, and the mean chromosome length are negatively correlated to the longitude (r = > 0.99, p < 0.01). This indicates that the populations that are distributed in higher longitude contained smaller chromosomes.
Moreover a significant positive correlation was observed between the number of chromosomes, total size of chromosome, the mean chromosome length and altitude (r = > 0.99, p < 0.01). This revealed that the populations that are distributed in higher altitude had lower value of total chromosome number and size.
Different clustering methods and ordination of the Chara species and varieties based on karyotype data produced similar results; therefore, only the UPGMA dendrogram is presented and discussed. The UPGMA dendrogram (Figure ) produced two major clusters. The first major cluster contains two sub-clusters formed by the n = 14 chromosomes species. The first sub-cluster contained C. gymnophylla var. rohleana, C. gymnophylla and C. connivens, while the second sub-cluster was formed by C. vulgaris var. vulgaris, C. vulgaris var. longibracteata, C. braunii and C. contraria. The two sub-clusters were joined with some distance.
Figure 3. UPGMA cluster analysis of n = 14 and n = 28 Chara species and varieties based on mitotic data. vul1, vul2, vul3, vul4, vul5, vul6, vul7, vul8, vul9, vul10, vul11 and vul12 are populations 1–12 of C. vulgaris var. vulgaris; long1, long2, long3 and long4 are populations 1–4 of C. vulgaris var. longibracteata; rohl1, rohl2, rohl3, rohl4, rohl5, rohl6 and rohl7 are populations 1–7 of C. gymnophylla var. rohlenae; gymn1, gymn2, gymn3, gymn4, gymn5 and gymn6 are populations 1–6 of C. gymnophylla; brau1 and brau2 are populations 1 and 2 of C. braunii; cont1, cont2 and cont3 are populations 1–3 of C. contraria; con is the population of C. connivens; glob1 is the population of C. globate (respectively in accordance with Table ).
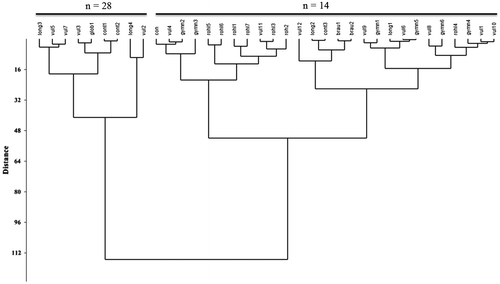
The species and varieties with n = 28 haploid chromosome numbers (two varieties of C. vulgaris, C. contraris and C. globate) formed the second main cluster and were placed far from the first major cluster.
Discussion
The results obtained indicate the occurrence of two different chromosome numbers of n = 14 and n = 28 in C. contraria, C. vulgaris and C. gymnophylla. The earlier studies on C. vulgaris (Sato Citation1961; Guerlesquin Citation1967a, Citation1967b; Mirasidov Citation1971; Grant and Proctor Citation1972; Khatun et al. Citation2009) reported the occurrence of n =14, 18, 28, and 42, but n = 14 and 28 are the most frequent chromosome counts for this species (Singh and Chaudhary Citation2010).
Polyploidy and genome duplication are the two major evolutionary phenomena that play prominent roles in speciation within charophytes (Casanova Citation2015). Karyological changes are known to be strong within a single species complex as well. For example, Wood and Imahori (Citation1965) identified eight varieties within the C. vulgaris complex, of which one variety (C. vulgaris var. vulgaris) had 22 forms. In this variety only seven forms have been investigated cytologically so far and all showed karyotype differences (Guerlesquin Citation1964, Citation1967; Mirasidov Citation1971; Bhattacharya Citation1972; Peshkov et al. Citation1974; Noor and Mukherjee Citation1977; Ramjee and Bhatnagar Citation1978). This study is the first to report the chromosome number in C. vulgaris var. vulgaris and C. vulgaris var. longibracteata.
In the present study, the Chara species and varieties studied differed in their karyotype formulae, indicating changes in the form of their chromosomes possibly due to structural changes such as inversion and translocation. These are qualitative changes in the genome (Sheidai and Rashid Citation2007).
ANOVA and LSD tests revealed significant differences in the total size of chromosomes, size of the long arms, size of the short arms and the arm ratio among the populations of each species and varieties studied, indicating the role of quantitative chromatin material (DNA) changes in the Chara species diversification.
The karyotype within the genus Chara is considered to be symmetrical and the majority of taxa have a dominance of metacentric and sub-metacentric chromosomes (Singh and Chaudhary Citation2010; Khatun et al. Citation2009). In present study, four Chara species showed high values of TF% (> 40) and contained a symmetrical karyotype. They were placed in 1A, 2A, 1B and 2B classes of the Stebbins system. Therefore, our findings confirmed the earlier belief of primitive karyotype status in the Charophytes (Mukhopadhyay and Ray Citation1995).
We observed that the CVCL versus MCA diagram is preferable to scatter diagrams based on other indices. It is suggested that this diagram is better suited to demonstrate karyotype relationships among taxa, especially when chromosome size variation is negligible (Peruzzi and Eroğlu Citation2013).
The cluster analysis of the joint n = 14 and n = 28 species and varieties, which was based on relative data, separated these species in two different groups. This result indicated that karyological features in species with different chromosome number are differentiated from each other. As our observation revealed, some of these features may be correlated with geographical features. For example, at higher longitudes, Chara species had higher chromosome number and a lower value of total chromosome size.
Many Chara species show morphological variability possibly under the influence of environmental conditions (Wood and Imahori Citation1965; Caisová and Gabka Citation2009). The present study reveals karyological features of some Chara species that may be responsible for Chara species delimitations. This hypothesis may be investigated in further studies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgments
The authors thank anonymous reviewers for valuable comments on an earlier draft.
References
- Ahmadi A, Riahi H, Sheidai M, Raam JV. 2012. Some Charophytes (Characeae, Charophyta) from Central and Western of Iran including Chara kohrangiana species nova. Cryptogamie, Algol. 33(4):359–390.
- Bhattacharya SS. 1972. Cytological studies on some Libyan charophytes. Libyan J Sci. 2:1–7.
- Bold HC, Wynne MJ. 1985. Introduction to the algae: Structure and reproduction. Englewood Cliffs, NJ: Prentice-Hall.
- Caisová L, Gąbka M. 2009. Charophytes (Characeae, Charophyta) in the Czech Republic: taxonomy, autecology and distribution. Fottea. 9(1):1–43.
- Casanova MT. 2005. An overview of Chara in Australia (Characeae, Charophyta). Aust Syst Bot. 18:25–39.
- Casanova MT. 2015. Chromosome numbers in Australian charophytes (Characeae, Charophyceae). Phycologia. 54(2):149–160.
- Casanova MT, Karol KG. 2014. A revision of Chara sect. Protochara, comb. et stat. nov. (Characeae: Charophyceae). Aust Syst Bot. 27:23–37. doi:10.1071/SB13016.
- Chatterjee P. 1976. Cytotaxonomical studies of West Bengal charophyta: karyotype analysis in Chara Braunii. Hydrobiologia. 49(2):171–174.
- Corillion R. 1975. Flore des Charophytes (Characées) du Massif armoricain. In: des Abbayes H, Claustres G, Corillion R, Dupont P, editors. Flore et végétation du Massif Armoricain. Paris: Jouve Éditeurs. IV, 216 pp.
- Gillet C. 1959. Nombres chromosomiques de plusieurs especes de charophycees (genres Nitella et Chara). Rev Cytol Biol Veg. 20:229–234.
- Grant MC, Proctor VW. 1972. Chara vulgaris and Chara contraria: patterns of reproductive isolation for two cosmopolitan species complexes. Evolution. 26:267–281.
- Guerlesquin M. 1964. Contribution a l’etude chromosomique des Charophycees d’Italie peninsulaire. Rev gen Bot. 71:283–292.
- Guerlesquin M. 1967a. Recherches caryotypiqoes et cytotaxonomiques chez les Charophycees d’Europe occidentale et d’Afrique du Nord. Bull Soc Sci Bretag. 41:1–265.
- Guerlesquin M. 1967b. Recherches caryotypiques et cytotaxinomiques chez les charophycees (PhD Thesis presented to the Faculty of Toulouse University of Sciences, Angers, France). Paris: Jouve Publishers.
- Guerlesquin M. 1984. Nombres chromosomiques et plöidie chez les Charophytes. Cryptogamie, Algologie. 23:115–126.
- Hammer Ø, Harper DAT, Ryan PD. 2013. PAST: Paleontological Statistics software package for education and data analysis. Palaeont Electron. 4(1):9.
- Khatun W, Ud-Deen MM, Kabir G. 2009. Karyotype of some species of Chara and Nitella (Charophyta) from Bangladesh. Bangl J Bot. 38:205–207.
- Krause W. 1997. Charales (Charophyceae). Süsswasserflora von Mitteleuropa. Band 18. Jena: Gustav Fischer Verlag. 202 pp.
- Levan AK, Fredga K, Sandberg AA. 1964. Nomenclature for centromic position on chromosomes. Hereditas. 52:201–220.
- Mirasidov TI. 1971. Nombre chromosomique de quelques especes de Characees d Ouzbekistan (in Russian). Uzbekbiol Zh. 15:59.
- Mukhopadhyay A, Ray S. 1995. Karyology and cytotaxonomy of Chara wallichii and Chara braunii from West Bengal. India. Acta Bot Gall: Bot Lett. 142(7):787–791.
- Noor MN, Mukherjee S. 1977. Some new records of chromosome numbers in Indian Charophyta. Cytologia. 42:227–232.
- Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Pl Syst Evol. 258:39–48.
- Peruzzi L, Eroğlu H. 2013. Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogen. 7(1):1–9.
- Peshkov MA, Mirsaidov TJ, Tikhomirova LA. 1974. Study of chromosome numbers and tentative karyotypes of some charophyte algae of the Uzbekistan SSR. Izvestia Akad Nauk SSSR Ser Biol. 5:672–680.
- Podani J. 2000. Introduction to the exploration of multivariate biological data. Leiden: Backhuys; p. 407.
- Proctor VW. 1971. Chara globularis Thuillier (= C. fragilis Desvaux): breeding patterns within a cosmopolitan complex. Limnol Oceanogr. 16:422–436.
- Ramjee, Bhatnagar SK. 1978. Studies on Charophyta from Rohilkhand Division I. Moradabad, taxonomic enumeration and chromosome counts. Phykos. 17:87–92.
- Romero Zarco C. 1986. A new method for estimating karyotype asymmetry. Taxon. 35:526–530.
- Sarma YSRK, Khan M. 1965. A preliminary report on the survey of chromosome numbers of Indian Charophyta. Nucleus. 8:33–38.
- Sarma YSRK, Ramjee. 1971. Significance of chromosome numbers in charophyta, a discussion. Caryologia. 24(4):391–401.
- Sato D. 1961. The protokaryotype and phylogeny in plants. Sci Pap Coll Gen Educ Univ Tokyo. 9(2):303–325.
- Sawa T. 1965. Cytotaxonomy of the Characeae: karyotype analysis of Nitella opaca and Nitella flexilis. Am J Bot. 52(9):962–970.
- Sheidai M, Arman M, Zehzad B. 2002. Chromosome pairing and B-chromosomes in some Aegilops species and populations of Iran. Caryologia. 55:261–271.
- Sheidai M, Jalilian N. 2008. Karyotypic studies in some species and populations of the genus Lotus L. in Iran. Acta Bot Croat. 67:45–52.
- Sheidai M, Maasoumii AR, Pakravan M. 1996. Karyotypes of some Astragalus taxa (Sec. Xiphidium Bge) from Iran. Nucleus. 39(3):111–113.
- Sheidai M, Rashid S. 2007. Cytogenetic study of some Hordeum L. species in Iran. Acta Biol Zseged. 51:107–112.
- Singh KV, Chaudhary BR. 2010. The first cytotaxonomic report in two forms of Chara vulgaris complex (Div. Charophyta). Res Env. Life Sci. 3(3):113–114.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold. 216 p.
- Watanabe K, Yahara T, Denda T, Kosuge K. 1999. Chromosomal evolution in the genus Brachyscome Asteraceae, Astereae): statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. J Plant Res. 112:145–161.
- Wood RD, Imahori K. 1965. Monograph of the Characeae Vol. 1 & 2. Weinheim, Germany: Von J. Cramer. 904 p.
