Abstract
Most chromosome preparation techniques use colchicine or colcemid in order to arrest cell division during metaphase, which usually involves the use of high cost materials. This research was aimed to simplify fish chromosome preparation technique without using colchicine or colcemid. Ten fish of different ages, namely one-, two- and three-month-old Nile tilapias, were used in this research. Tissues were collected on the caudal fin of the fish. Chromosome spreads was easily observed, using Giemsa or silver staining, five hours after the whole preparation process. The results showed that the one- and two-month-old fish had better mitotic index (0.008 and 0.01, respectively) compared to those of the three-month-old fish (0.005). This method is rapid, cost-efficient and applicable for ploidy level determination, and also keeps fish alive for subsequent evaluations.
Introduction
Chromosome set manipulation is an important practice for aquaculture purposes (Khan et al. Citation2000; Benfey Citation2001). One of the chromosome set manipulation techniques that is being currently studied is polyploidization. Ploidy level determination plays a crucial role in ensuring the success of chromosome set manipulation related activities, especially polyploidization. A fast and accurate method of ploidy level determination is necessary to evaluate the success of polyploidization at earlier life stages, as the treatment does not always yield 100% target population.
Factors such as the equipment, the experience of the laboratory staff, and the handling species and their life stage and size strongly influence the ploidy determination method. A combination of several simple techniques could greatly help speed and accuracy in determining ploidy levels (Jankun et al. Citation2007). Some ploidy identification methods have been presented (Tave Citation1993; Boron Citation1994), including chromosome counting (Benfey Citation1999), erythrocyte size examination (Woznicki and Kuzminski Citation2002; Ocalewicz et al. Citation2007; Dewi et al. Citation2010), DNA content measurement (Harrella et al. Citation1998) and nucleoli counting (Carman et al. Citation1991, Citation1992, Citation1997; Carman Citation1992; Mukti et al. Citation2001; Mukti Citation2005; Mukti and Mubarak Citation2007).
Among the mentioned methods, chromosome counting is the most accurate one, because of its ploidy level representing its chromosome set number. The other methods usually represent an indirect relationship between ploidy level and biological characters as predictor. Furthermore, chromosome counting depends on a good chromosome preparation method.
Generally, the chromosome spread method is not easily applied to large number of small sized fish. In most fish chromosome preparation techniques, tissues are collected from different parts of the fish body, such as kidney (Blanco et al. Citation2010; Poletto et al. Citation2010; Martinez et al. Citation2011; Liu et al. Citation2012), liver (Affonso and Galetti Citation2005), spleen (Affonso and Galetti Citation2005), intestine (Kligerman and Bloom Citation1977), gill epithelium (Hashimoto et al. Citation2009; Liu et al. Citation2012), whole body (Shao et al. Citation2010; Pradeep et al. Citation2011), blood (Gold et al. Citation1990; Eugenio et al. Citation1993; Jacobina et al. Citation2011), skin fibroblasts (Tantithakura et al. Citation1993), bone marrow (Manosroi et al. Citation2003), gonads (Liu et al. Citation2012) and fin epithelium (Fontana et al. Citation2008). The above preparation techniques involved the use of metaphase arresting agents, such as colchicine or colcemid. These agents present some disadvantages such longer preparation time, fish death and cost (especially in developing countries). Chemicals such as colchicine or colcemid are known as carcinogens and toxic compounds.
Thus, developing a fast and simple fish chromosome preparation technique is greatly important in assessing ploidy level in fish. This research was aimed to simplify fish chromosome preparation techniques without using colchicine or colcemid.
Materials and methods
The experiment was conducted in the Laboratory of Fish Breeding and Genetics, Aquaculture Department, Faculty of Fisheries and Marine Sciences, Bogor Agricultural University, Bogor, Indonesia. Nile tilapia at three different ages (one, two and three months) produced through artificial fertilization were used for chromosome preparations in the current research. Ten diploid fish were collected from each age group and used for chromosome preparation without using colchicine or colcemid. Ten one-month-old triploid fish that were used for chromosome preparation were obtained using heat shock treatment at 41°C for 4 min, 4 min after fertilization. Chromosome preparation technique of 10 one-month-old fish, both diploid and triploid with colchicine-immersed treatment (0.007% w/v) for seven hours were used as control.
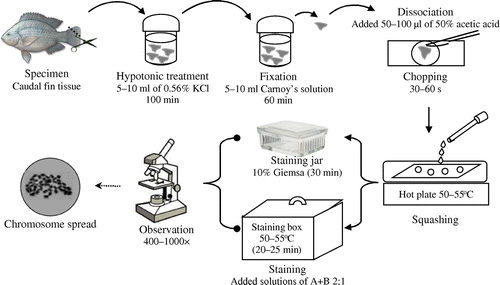
The solid tissue method (Kligerman and Bloom Citation1977) and staining method (Howell and Black Citation1980) were used as standard chromosome preparation and staining protocols, respectively. The caudal fin tissues were collected by quickly cutting 0.25 cm2 pieces (without anesthetic) and immediately transferred into 5–10 ml hypotonic solution (0.56% KCl) and placed into an incubator at 29–30°C for 30, 40, 50 and 60 min (twice, respectively). Then, the tissues were fixed in 5–10 ml fresh and cool (7–10°C) Carnoy’s solution that consisted of absolute ethanol and acetic acid (3:1) for 30 min (twice). After the fixation process, epithelium cells in tissues were dissociated by adding 3–4 drops of 50% acetic acid and gently chopped using a sharp scalpel on concave glass object for 30–60 s until a cloudy cell suspension was formed. When dealing with a large number of samples, after fixation, the tissues were placed in Carnoy’s solution and stored in a refrigerator at 4°C for at most two weeks before the next process.
By using a 100 μl chip micropipette, the cell suspension was gently pipetted and squashed on a very clean and warm slide glass that was placed on hot plate (50–55°C). This concludes from our experiences dealing with fish chromosome preparation technique for long time, especially it's correlated with availability of slide glass in our country, although we usually use new slide glasses but for chromosome preparation we need this step, as we prove under microscope that the new slide glasses often contain unwanted attached debris, to ensure the best result, it is recommended to immerse newly slide glasses in 96% ethanol for at least two hours before use in order to ensure their cleanliness. Squashing involved the following steps: after pipetting, all of the cell suspension was sucked from the concave glass object and dropped on a warm slide glass from a height of 4–5 cm and immediately sucked back to form 1–1.5 cm diameter rings (three rings per slide glass). It is recommended to make three preparates for each sample. This case also comes from our experiences, that sometime we did some troubles at the next steps (e.g. staining process). By doing so, we can save important sample slides without repeat a long process of preparation from the beginning. After the air-drying process for 2–3 minutes, the preparates were stained.
Preparates were stained using Giemsa stain (Kligerman and Bloom Citation1977) or silver stain (Howell and Black Citation1980). Giemsa staining was carried out by immersing the preparates in a freshly prepared 10% Giemsa solution in a phosphate buffer solution (PBS) at a pH of 6.9 for 30 min. Meanwhile, silver staining was carried out by the following procedure: two drops of a solution A (made by dissolving 10 g of AgNO3 in 20 ml distilled water) and one drop of a solution B (made by dissolving 0.2 g of gelatin in 5 ml warm 50% glycerin solution and adding one drop of formic acid) were dropped on the preparates, gently mixed and spread using a toothpick without scratching the cell film laid on the preparates. They were then placed into a high humidity staining box at 50–55°C for 20–25 min.
Both Giemsa and silver stained preparates were carefully rinsed using distilled water and air dried. Completely dried preparates were observed under 400 × and 1000 × magnifications using a BH2-RFCA Olympus microscope (Olympus Optical Ltd. Shinjuku-ku,Tokyo, Japan), which was equipped with a camera. Mitotic index of different fish age groups (three preparates, respectively) were determined by calculating the ratio of chromosome spreads (prophase, metaphase, anaphase and telophase) to the total number of 53 × 103–59 × 103 interphase cells.
Data of mitotic index were statistically analyzed using ANOVA with Minitab 16 software (https://www.minitab.com/en-us/products/minitab/) and a Tukey’s test with a confidence level of 95%. The whole chromosome preparation procedure is represented in Figure .
Results
The results of this study showed that chromosome spreads were observed in all preparates of different fish age groups (one-, two- and three-month-old). Microscope observation at 400× magnification showed 3–4 chromosome spreads that were observed in a view field (Figure ). The younger tilapia, one and two months old, showed more chromosome spreads compared to the three-month-old tilapia. Mitotic index of the one- and two-month-old fish groups (Table ) were 0.008 and 0.01 respectively, and were significantly higher (p < 0.05) compared to the three-month-old fish group (0.005).
Figure 2. Chromosome spreads (arrows) observed at 400 × magnification; prepared without (a) and with (b) metaphase arrest treatments.
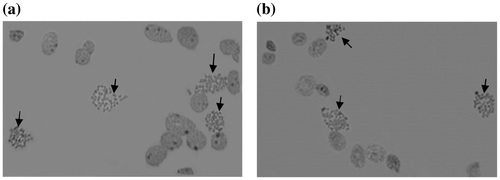
Table 1. Chromosome spreads and mitotic index of different fish age groups in tilapia.
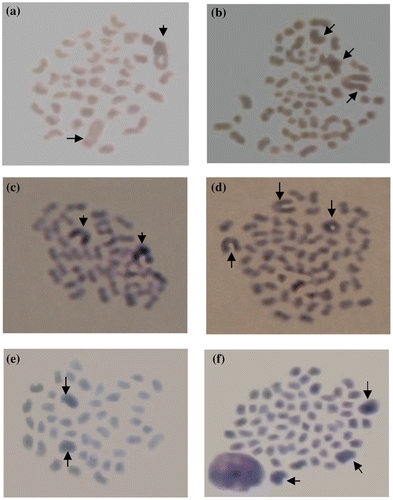
The chromosome spreads of diploid and triploid fish were easily distinguished using either Giemsa or silver stains (Figure ). A better chromosome spread was observed in the Giemsa staining process. No significant difference was observed in term of chromosome spreads between treatments without or with metaphase arresting agents.
Figure 3. Silver stained diploid (a) and triploid (b) chromosomes and Giemsa stained diploid (c) and triploid (d) chromosomes prepared without metaphase arrest treatment compared with Giemsa stained diploid (e) and triploid (f) chromosomes prepared with metaphase arrest treatment (arrows indicate giant chromosomes).
Discussion
Mitotic index is an important factor in successfully obtaining a high number of chromosome spreads in the chromosome preparation techniques. The higher the mitotic index, the higher is the opportunity to obtain a chromosome spread. In the current study, one-, two- and three-month-old fish were reliable as specimens in the chromosome preparation without using colchicine or colcemid. They showed relatively good chromosome spreads for ploidy level determination in tilapia. The one- and two-month-old fish showed higher mitotic index compared to the three-month-old fish. This might be due to the fact that younger fish have a high cell division rate due to their development and high growth rate. Conversely, the three-month-old fish showed low mitotic index, possibly because they have begun the slow growth phase, in which their gonadal development actively starts. During that phase, metabolic energy for somatic growth begins to decrease due to its allocation to gonadal development, initiating the reproduction phase. This phenomenon is called sigmoid growth pattern, in which young fish grow rapidly, but gradually slow as they get older. Furthermore, caudal fin tissue used in this research might have a fast cell degeneration process, as a consequence of its direct exposure to various environmental factors, making it reliable to be used for chromosome preparation (Fontana et al. Citation2008).
Chromosome preparation techniques are relatively difficult in teleost fish compared to other vertebrates, such as mammals. One of the main difficulties is getting a high quality metaphase spread (Esmaily and Kalbasi Citation2004), especially for ploidy level determination. Simplifying the chromosome preparation techniques in fish, to provide a high number of metaphase spreads for ploidy level determination, is of great importance. One chromosome preparation technique involves air-drying using solid tissue as the source in the preparation process. Ganai et al. (Citation2011) reported that the air-drying technique was originally developed for mammals, but is being widely used in fish chromosome preparation. The chromosome preparation method that was used in this research was faster than most methods that have been used so far. This is due to the absence of metaphase arresting agent, such as colchicine and colcemid, compared to conventional methods that require a longer period of time. The conventional fish chromosome preparation technique using colchicine or colcemid is done through immersion or injection and requires 5–10 h for the first step of the preparation process alone. In the current study, the non-use of colchicine or colcemid saved about 5–10 h in the preparation process and the ploidy level was quickly determined; the whole process took about five hours. It is an effective time saving technique, especially for chromosome preparation routine work, when a large number of samples should be evaluated. In addition to time efficiency, a considerable cost was saved by not using metaphase arresting agents such as colchicine or colcemid, which are relatively limited and expensive.
Chromosomes of Nile tilapia are relatively easy to observe and confirm, because of the uniqueness of their marker homologues, the giant chromosomes (GC), which are characterized as larger sized chromosomes compared to others, such as haploid, diploid, triploid and tetraploid that bear 1, 2, 3 and 4 GCs, respectively. In the ploidy level determination, the GC was very helpful in validating the ploidy level, especially when aneuploid chromosome spread was observed.
Although this method has proved to successfully identify the ploidy level of Nile tilapia, the success might be correlated to proper handling of the step by step protocol, i.e. fish handling before tissue collection, hypotonic treatment, tissue fixation, chopping, squashing and staining. In the current research, hypotonic treatment significantly influenced the result, especially in terms of numerous, good and easily countable chromosome spreads. Mishandling one of these steps might lead to imperfect chromosome spread and appearance due to chromosome overlapping. Pradeep et al. (Citation2011) reported that hypotonic treatment is an important factor in improving the chromosomes spread. Hypotonic treatment allows cell swelling that causes chromosome disruption and dispersion, when the cell contents scattered on the slide. In the current study it was observed that the optimum condition for hypotonic treatment was using a 0.56% KCl solution at 29–30°C for 100 min.
This technique might be effectively used in determining ploidy level in fish species with a relatively low number of chromosomes, such as Xiphophorus spp., 2n = 48 (Tantithakura et al. Citation1993), flounder, 2n = 44–50 (Winkler et al. Citation2004; Fujiwara et al. Citation2007), Barbodes spp., 2n = 48 (Defira Citation2004), Characidium spp., 2n = 50 (CitationPansonato-Alves et al. 2011), bitterlings, 2n = 46 (Ueda et al. Citation2001), goby, 2n = 46 (Esmaily and Kalbasi Citation2004), loach, 2n = 50 (Vasilľev and Vasiľeva Citation2008) and Clarias spp., 2n = 54 (Na-Nakorn and Lakhaanantakun Citation1993). However, further research is needed for fish species with a large number of chromosomes, such as carp, which has more than 150 (Zhou and Gui Citation2002). The GC number was correlated with ploidy level in tilapia. Using this marker, ploidy level determination was done by simply counting the GC number instead of counting the chromosome number in a given spread. However, this technique is not recommended for karyotyping studies, since morphological details were not easily observed in homologue chromosomes.
Another advantage of this method is that the fish were not sacrificed, which might allow other analyses to be conducted on the polyploid target fish, such as physiology, behavioral and other genetic aspects. This chromosome preparation without metaphase arresting agent is a promising technique that could be applied to a variety of fish species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding information
This work was supported by The Ministry of Research, Technology and Higher Education, Republic of Indonesia sponsored this research by BPP-DN scholarship.
Acknowledgements
The authors would like to thank the Ministry of Research, Technology and Higher Education, Republic of Indonesia, which has sponsored this research through BPP-DN scholarship. The authors would also like to thank the deceased Prof. Komar Sumantadinata, who has provided tremendous guidance and support. The authors appreciate the comments and corrections given by a reviewer and proof reader, which improved this article.
References
- Affonso PRAM, Galetti PM Jr. 2005. Chromosomal diversification of reef fishes from genus Centropyge (Perciformes, Pomacanthidae). Genetica. 123(3):227–233.
- Benfey TJ. 1999. The physiology and behaviour of triploid fishes. Rev Fish Sci. 7(1):39–67.
- Benfey TJ. 2001. Use of sterile triploid Atlantic salmon (Salmo salar L.) for aquaculture in New Brunswick. Canada. ICES J Mar Sci. 58(2):525–529.
- Blanco DR, Lui RL, Bertollo LAC, Diniz D, Filho OM. 2010. Characterization of invasive fish species in a river transposition region: evolutionary chromosome studies in the genus Hoplias (Characiformes, Erythrinidae). Rev Fish Biol Fish. 20(1):1–8.
- Boron A. 1994. Use of erythrocyte measurements to detect natural triploids of spined loach Cobitis taenia (L.). Cytobios. 78(315):197–202.
- Carman O. 1992. Chromosome set manipulation in some warm water fish (Doctoral Thesis). Tokyo, Japan: Tokyo University of Fisheries 131 pp.
- Carman O, Alimuddin, Sastrawibawa S, Arfah H. 1997. Determination of sex chromosome and nucleoli in red tilapia fish (Oreochromis sp.). Zuriat. 8(2):83–89. (in Indonesian with English abstract)
- Carman O, Oshiro T, Takashima F. 1991. Estimation of effective condition for induction of triploidy in goldfish, Carassius auratus Linnaeus. J Tokyo Univ Fish. 78(2):127–135.
- Carman O, Oshiro T, Takashima F. 1992. Variation in the maximum number of nucleoli in diploid and triploid common carp. Nippon Suisan Gakkaishi. Bull Japan Soc Sci Fish. 58(12):2303–2309.
- Defira CN. 2004. Morphological variation, karyotype and isozyme pattern of lalawak (Barbodes balleroides) and lalawak jengkol (Barbodes sp.) from Cikandung River and ponds at Buah Dua, Sumedang (Master Thesis). Graduate School, Bogor Agricultural University, Bogor, Indonesia. 84 pp. (in Indonesian with English abstract)
- Dewi IS, Mukti AT, Mubarak AS. 2010. Analysis of volume and erythrocyte cell number of tetraploidization-treated Nile tilapia (Oreochromis niloticus). Proceedings of International Seminar on From Ocean for Food Security, Energy, and Sustainable Resources and Environment, Nov 18, 2009, Airlangga University, Surabaya, Indonesia. pp. 97–100.
- Esmaily A, Kalbasi M. 2004. Karyological study on bighead gobie (Neogobius kessleri) in Mahmoudabad Area (South Caspian Sea). Proceedings of the Fourth International Iran and Russia Conference on Agriculture and Natural Resources. Sept 8–10, 2004, Shahrekord, Iran. pp. 1434–1441.
- Eugenio MRN, Samonte RV, Galman O. 1993. Optimization of conditions for tilapia whole blood culture for chromosome studies. In: Dodson JJ, Soewardi K, Phang VPE, Enriques GL, Na-Nakorn U, Sukimin S, editors. Symposium on Fish Genetics and its Application to Aquaculture and Fishery Management. Seameo Biotrop: Bogor, Indonesia; p. 41–44.
- Fontana F, Lanfredi M, Kirschbaum F, Garrido-Ramos MA, Robles F, Forlani A, Congiu L. 2008. Comparison of karyotypes of Acipenser oxyrinchus and A. sturio by chromosome banding and fluorescent in situ hybridization. Genetica. 132(3):281–286.
- Fujiwara A, Fujiwara M, Nishida-Umehara C, Abe S, Masaoka T. 2007. Characterization of Japanese flounder karyotype by chromosome bandings and fluorescence in situ hybridization with DNA markers. Genetica. 131(3):267–274.
- Ganai FA, Yousuf AR, Tripathi NK, Zargar UR. 2011. On the chromosomes of two cyprinid fishes of the subfamily Schizothoracinae from Kashmir. Nature Sci. 9(3):53–61.
- Gold JR, Li YC, Shipley NS, Powers PK. 1990. Improved methods for working with fish chromosomes with a review of metaphase chromosome banding. J Fish Biol. 37(4):563–575.
- Harrella RM, Van Heukelema W, Kerbyb JH. 1998. A comparison of triploid induction validation techniques. Progress Fish-Cultur. 60(3):221–226.
- Hashimoto DT, Laudicina A, Bortolozzi J, Foresti F, Porto-Foresti F. 2009. Chromosomal features of nucleolar dominance in hybrids between the Neotropical fish Leporinus macrocephalus and Leporinus elongatus (Characiformes, Anostomidae). Genetica. 137(2):135–140.
- Howell WM, Black DA. 1980. Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 36(8):1014–1015.
- Jacobina UP, Cioffi MB, Souza LGR, Calado LL, Tavares M, Manzella J Jr, Bertollo LAC, Molina WF. 2011. Chromosome mapping of repetitive sequences in Rachycentron canadum (Perciformes: Rachycentridae): Implications for karyotypic evolution and perspectives for biotechnological uses. J Biomed Biotechnol. 218231. doi: 10.1155/2011/218231.
- Jankun M, Kuzminski H, Furgala-Selezniow G. 2007. Cytologic ploidy determination in fish – an example of two salmonid species. Env Biotechnol. 3(2):52–56.
- Khan TA, Bhise MP, Lakra WS. 2000. Chromosome manipulation in fish – a review. Ind J Anim Sci. 70(2):213–221.
- Kligerman AD, Bloom SE. 1977. Rapid chromosome preparation from solid tissues of fish. J Fish Res Board Can. 34(2):266–269.
- Liu S, Hui TH, Tan SL, Hong Y. 2012. Chromosome evolution and genome miniaturization in minifish. PLoS ONE. 7(5):1–7.
- Manosroi J, Petchjul K, Mevatee U, Manosroi A. 2003. Karyotype analysis of the hybrid, Thai red tilapia (Oreochromis niloticus Linn. X Oreochromis mossambicus Linn). J Biol Sci. 3:612–617.
- Martinez PA, Jacobina UP, Molina WF. 2011. Comparative cytogenetics and heterochromatic patterns in two species of the genus Acanthostracion (Ostraciidae: Tetraodontiformes). Mar Genom. 4(3):215–220.
- Mukti AT. 2005. Difference of success polyploidization level of common carp (Cyprinus carpio Linn.) by heat shock. J Biol Res. 10(2):133–138 (in Indonesian with English abstract).
- Mukti AT, Mubarak AS. 2007. Identification of nucleoli variation in common carp (Cyprinus carpio) with different ploidy level. Proceedings of the National Seminar on Breeding, Genetics, and Biotechnology of Fisheries; Nov 12, 2007; Bali, Indonesia. pp. 80–83. (in Indonesian with English abstract)
- Mukti AT, Rustidja, Sumitro SB, Djati MS. 2001. Polyploidization of common carp (Cyprinus carpio L.). Biosain J Life Sci. 1(1):111–123. (in Indonesian with English abstract)
- Na-Nakorn U, Lakhaanantakun A. 1993. Comparison between the performance if diploid and triploid Clarias macrocephalus. In: Dodson JJ, Soewardi K, Phang VPE, Enriques GL, Na-Nakorn U, Sukimin S, editors. Symposium on Fish Genetics and its Application to Aquaculture and Fishery Management. Seameo Biotrop: Bogor, Indonesia; p. 79–86.
- Ocalewicz K, Jankun M, Luczynski M. 2007. Chromosome set manipulations and cytogenetic characteristic of polyploid, gynogenetic, and androgenetic fish strains. In: Pisano E, Ozouf-Costaz C, Foresti F, Kapoor BG, editors. Fish Cytogenetics. New Hampshire, USA: Science Publishers; p. 289–332.
- Pansonato-Alves JC, Oliveira C, Foresti F. 2011. Karyotypic conservatism in samples of Characidium cf. zebra (Teleostei, Characiformes, Crenuchidae): Physical mapping of ribosomal genes and natural triploidy. Genet Mol Biol. 34(2):208–213.
- Poletto AB, Ferreira IA, Cabral-de-Mello DC, Nakajima RT, Mazzuchelli J, Ribeiro HB, Venere PC, Nirchio M, Kocher TD, Martins C. 2010. Chromosomal differentiation patterns during cichlid fish evolution. BMC Genet. 11:50.
- Pradeep PJ, Srijaya TC, Zain RBM, Papini A, Chatterji AK. 2011. A simple technique for chromosome preparation from embryonic tissues of teleosts for ploidy verification. Caryologia. 64(2):235–241.
- Shao C-W, Wu P-F, Wang X-L, Tian Y-S, Chen S-L. 2010. Comparison of chromosome preparation methods for the different developmental stages of the half-smooth tongue sole, Cynoglossus semilaevis. Micron. 41(1):47–50.
- Tantithakura O, Andreas F, Anders A, Forester W. 1993. High resolution technique: A new approach to study fish chromosomes. In: Dodson JJ, Soewardi K, Phang VPE, Enriques GL, Na-Nakorn U, Sukimin S, editors. Symposium on Fish Genetics and its Application to Aquaculture and Fishery Management. Seameo Biotrop: Bogor, Indonesia; p. 45–53.
- Tave D. 1993. Genetics for fish hatchery managers. Connecticut: Avi Publishing.
- Ueda T, Naoi H, Arai R. 2001. Flexibility on the karyotype evolution in bitterlings (Pisces, Cyprinidae). Genetica. 111(1):423–432.
- Vasiľev VP, Vasiľeva ED. 2008. Comparative karyological analysis of mud loach and spined loach species (genera Misgurnus and Cobitis) from the Far East region of Russia. Folia Zoology. 57(1–2):51–59.
- Winkler FM, Garcia-Melys D, Palma-Rojas C. 2004. Karyotypes of three South East Pacific flounder species of the family Paralichthyidae. Aquacult Res. 35(13):1295–1298.
- Woznicki P, Kuzminski H. 2002. Chromosome number and erythrocyte nuclei length in triploid brook trout (Salvelinus fontinalis). Caryologia. 55(4):295–298.
- Zhou L, Gui J. 2002. Karyotypic diversity in polyploidy gibel carp, Carassius auratus gibelio Bloch. Genetica. 115(2):223–232.