Abstract
Karyotype structures in representative taxa of the genus Allium L. are compared, based on Feulgen-stained somatic metaphase chromosomes. The karyotypes of Allium arlgirdense, A. anacoleum and A. microspathum (in subg. Allium), and A. rhetoreanum and A. shirnakense (in subg. Melanocrommyum) are described for the first time. The karyological analyses indicate the relationships among the species with respect to asymmetry indices. All Allium taxa studied are diploid with 2n = 2x = 16 chromosomes. One B chromosome is detected in the chromosomes of A. microspathum. All chromosome sets of the studied taxa consist of seven median chromosomes and one submedian chromosomes and all submetacentric pairs bear a satellite on the short arms. The lengths of the chromosomes show remarkable differences particularly at the subgenus level. The three subgenus of genus Allium show a clear tendency to have karyotypes distinct on asymmetry grounds. Karyotype asymmetry relationships are discussed according to the bidimensional scatter plots of A1–A2 and CVCL–MCA.
Introduction
According to Koyuncu (Citation2012), the genus Allium L. is represented in Turkey by 196 taxa, many of which endemic (c.39%) to this territory. Chromosome numbers have been reported for approximately 75% of these taxa. Most of these taxa are diploid. However polyploidy has been reported some previous counts for Allium (Goldblatt and Johnson 1979–; Özhatay et al. Citation2000). A basic chromosome number of x = 8 dominates in most subgenera whereas nearly all taxa of subg. Amerallium are characterized by x = 7 chromosomes (Fritsch and Astanova Citation1998).
This study focuses on six taxa including Allium arlgirdense, A. anacoleum, A. microspathum (in subg. Allium), A. rhetoreanum, A. shirnakense (in subg. Melanocrommyum) and A. oreophilum (in subg. Poryphrason) (Friesen et al. Citation2006). These taxa are rare for Flora of Turkey owing to populations with a very restricted area of occupancy, but A. oreophilum is more widely distributed than the others (Kollmann Citation1984).
Material and methods
The plant materials used in this study are listed in Table . Voucher specimens have been deposited in the Herbarium of University of Yüzüncü Yıl (VANF), Van, Turkey.
Table 1. Localities and voucher specimens of Allium taxa examined in the present study.
For karyological observations, four to eight individuals were used for each taxon in this study. Mitotic metaphase cells of root tips were obtained from planted bulbs which were collected in natural habitats from Turkey. Root tips were pretreated in 0.002 M 8-hydroxyquinoline at 4°C overnight, washed with distilled water and fixed in Carnoy’s solution (3:1 absolute ethanol: glacial acetic acid) for a minimum of 1 h. The root tips were hydrolysed for 10 minutes in 1N HCl at 60°C, stained using the standard Feulgen technique and squashes were prepared.
Microphotographs of good quality metaphase plates were taken using an Olympus BX53 (Tokyo, Japan) microscope equipped with a high-resolution digital camera. The somatic chromosome number and karyotype details were studied in a minimum of five spread metaphase plates from different individuals; mean values were used for the analysis. Metaphase observations and chromosome measures were made using the image analysis systems KAMERAM (ARGENİT Microsystems, İstanbul, Turkey). The karyotype was determined using the above-mentioned software for the recognition of homologues, couple ordering, chromosome classification and karyotype formula based on centromere position (Levan et al. Citation1964). Karyotype symmetry was determined calculating both CVCI and CVCL parameters (Paszko Citation2006), together with the recent MCA index (Peruzzi and Eroğlu Citation2013). Karyotype asymmetry was determined calculating both A1 and A2 parameters (Romero Zarco Citation1986).
Results
All Allium taxa studied were diploid with 2n = 2x = 16 chromosomes, but A. microspathum has a B chromosome. It is fixed for all cells and individuals analysed in this study (Figure ). The ideograms and parameters obtained are based on the mean values presented in Figure . All chromosome sets of the studied taxa consist of seven median chromosomes and one submedian chromosome and all submetacentric pairs bear a satellite on the short arms. Although one of the metacentric pair bears a satellite on the short arms of A. rhetoreanum (Figure d, ).
Figure 1 Somatic chromosomes in the studied taxa. (a) Allium arlgirdense; (b) A. anacoleum; (c) A. microspathum; (d) A. rhetoreanum; (e) A. shirnakense; (f) A. oreophilum. Scale bars 10 μm.
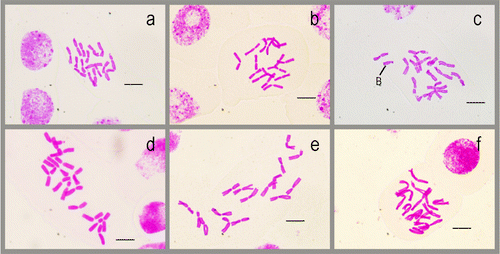
Figure 2 Haploid ideograms in the studied taxa. (a) Allium arlgirdense; (b) A. anacoleum; (c) A. microspathum; (d) A. rhetoreanum; (e) A. shirnakense; (f) A. oreophilum.
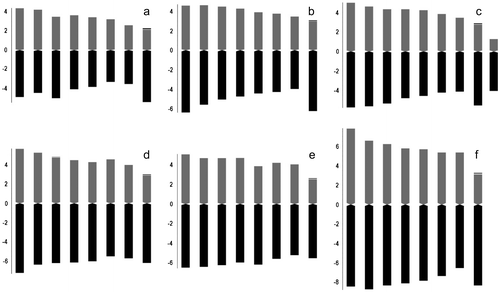
Chromosome measures, karyotype formulae and symmetry indices are given in Tables and for all investigated taxa.
Table 2. Karyotype formula according to Levan et al. (Citation1964) and characteristics of the investigated taxa.
Table 3. Karyo-morphometric parameters and symmetry indices for the investigated taxa.
Differences were not observed in the karyotype formulae of the studied taxa. However remarkable differences were identified in the lengths of the chromosomes, particularly at the subgenus level.
The ratio of shortest and longest chromosome ranged from 6.11 μm in A. arlgirdense (subg. Allium) to 16.26 μm in A. oreophilum (subg. Poryphrason). The ratio of shortest and longest haploid chromosome length ranged from 61.61 μm in A. arlgirdense (subg. Allium) to 109.73 μm in A. oreophilum (subg. Poryphrason). The smallest arm ratio was observed in A. arlgirdense (3.39) and the largest was observed in A. oreophilum (7.98) (Table ).
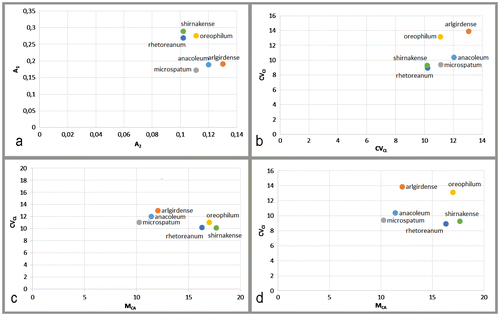
According to the intrachromosomal (A1) asymmetry indices, A. microspathum (0.17 ± 0.01) had the most symmetrical karyotype and A. oreophilum (0.29 ± 0.02) had the most asymmetrical karyotype among the Allium species analysed. According to values of A1, subg. Allium had the most symmetrical karyotype, then subg. Melanocrommyum and subg. Poryphrason (Table and Figure ).
Discussion
The current paper presents the somatic chromosome numbers and karyotypes of six rare taxa in three subgenera of the genus Allium distributed in the east Anatolian part of Turkey.
The chromosome number of A. oreophilum was reported as 2n = 16 in previous studies (Badr and Elkington Citation1977; Pogosian Citation1983; Hanelt et al. Citation1989; Özhatay Citation1993). Additionally for this species, Pogosian (Citation1983) reported two different chromosome numbers of 2n = 14, 16, whereas we found 2n = 2x = 16 as in other reports. The chromosome number, the karyotype formula and the ideogram of other five taxa were reported for the first time in this study.
The best way to represent karyotype asymmetry relationships among organisms is by means of bidimensional scatter plots (Romero Zarco 1986; Peruzzi et al. Citation2009). These scatter plots were generated with A1 and A2 (Romero Zarco Citation1986) and/or CVCI and CVCL (Paszko Citation2006; Peruzzi et al. Citation2009) parameters. A new parameter was added by Peruzzi and Eroğlu (Citation2013) which is called MCA (mean centromeric asymmetry).
According to the results of present investigation CVCL results positively correlated with MCA; CVCI did not correlate with CVCL and MCA. This evidence has endorsed the results of Peruzzi and Eroğlu (Citation2013).
As can be seen in Figure c, the three subgenus of genus Allium show a clear tendency to have karyotypes distinct on asymmetry grounds: subg. Allium, with clearly low intrachromosomal (MCA) and relatively high interchromosomal asymmetry (CVCL), subg. Melanocrommyum with higher intrachromosomal asymmetry and subg. Poryphrason with higher interchromosomal asymmetry. The results of a previous paper by the first author about the karyology of subg. Melanocrommyum showed values of A1 ranging from 0.23 to 0.39 (Genç et al. Citation2013), which endorses the results of the present investigation.
Subg. Poryphrason is close to subg. Melanocrommyum (Friesen et al. Citation2006). Therefore these subgenera are not distinct from each other.
In conclusion, according the results of this study, A1 and MCA parameters have separated species at the subgenus level for Allium.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Badr A, Elkington TT. 1977. Variation of Giemsa C-band and fluorochrome banded karyotypes, and relationships in Allium subgen. Molium. Plant Syst Evol. 128(1):23–35.
- Friesen N, Fritsch RM, Blattner FR. 2006. Phylogeny and new intrageneric classification of Allium L. (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso. 22(1):372–395.
- Fritsch RM, Astanova SB. 1998. Uniform karyotypes in different sections of Allium L. subgen. Melanocrommyum (Webb & Berth.) Rouy from Central Asia. Feddes Rep. 109(7-8):539–549.
- Genç İ, Özhatay N, Cevri M. 2013. A karyomorphological study of the genus Allium (sect. Melanocrommyum) from Turkey. Caryologia. 66(1):31–40.
- Goldblatt P, Johnson DE, editors. 1979. Index to plant chromosome numbers. St. Louis: Missouri Botanical Garden.
- Hanelt P, Fritsch RM, Kruse J, Maass H, Ohle H, Pistrick K. 1989. Allium L. sect. Porphyroprason Ekberg – Merkmale und systematische Stellung. Flora. 182(1-2):69–86.
- Kollmann F. 1984. Allium L. In: Davis PH, editor. Flora of Turkey and the East Aegaean Islands. Edinburgh: Edinburgh University Press. Volume 8; p. 98–211.
- Koyuncu M. 2012. Allium L. In: Güner A, Aslan S, Ekim T, Vural M, Babaç MT, editors. Türkiye Bitkileri Listesi (Damarlı Bitkiler). İstanbul: Nezahat Gökyiğit Botanik Bahçesi ve Flora Araştırmaları Derneği Yayını; p. 30–44.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52(2):201–220.
- Özhatay N. 1993. Allium in Turkey: distribution, diversity, endemism and chromosome number. In Demiriz H, Özhatay N, editors. 5th OPTIMA Meeting; 1986 Sep 8-15; İstanbul, Turkey. Istanbul University Press. p. 247–271.
- Özhatay N, Sadıkoğlu N, Johnson MAT. (2000). Index to Turkish Plant Chromosome Numbers. Allium L. In: Güner A, Özhatay N, Ekim T, Başer KHC, editors. Flora of Turkey and the east Aegean Islands. Edinburgh: Edinburgh University Press. Volume 11 Second Supplement; p. 445–460.
- Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 258(1-2):39–48.
- Peruzzi L, Eroğlu HE. 2013. Karyotype asymmetry: again, how to measure and what to measure? Compar Cytogenet. 7(1):1–9.
- Peruzzi L, Leitch IJ, Caparelli KF. 2009. Chromosome diversity and evolution in Liliaceae. Ann Bot. 103(3):459–475.
- Pogosian AI. 1983. Chromosome numbers of some species of the Allium (Alliaceae) distributed in Armenia and Iran. Bot Z. 68(5):652–660 (In Russian).
- Romero Zarco C. 1986. A new method for estimating karyotype asymmetry. Taxon. 35(3):526–530.