Abstract
The house mouse, Mus musculus, is known for its highly conserved morphology and chromosomal structure, but some chromosomal characters have been recognized as accurate taxonomic markers in this species, e.g. centromeric heterochromatin, which is useful to identify the taxonomic status of house mouse. In this study, comprehensive cytogenetic surveys have been carried out in five to eighteen specimens of house mouse from each locality of Iran using centromeric heterochromatin banding (C-banding). C-banding was performed on the karyotype, and the centromeric heterochromatin region size (CHRs) was calculated using karyotype analysis software. Results indicated that intra- and inter- population variation of C-band positive heterochromatin ranged from very large CHRs to small CHRs. Generally, chromosomes 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16 and 19 showed high variability in their CHRs whereas chromosomes 1, 2, 3, 13, 17, 18 and X had low variability in CHRs. Chromosome 4 had the largest CHRs in all populations and chromosomes 15, 17 and 19 showed the smallest CHRs. The entire Y-chromosome was C-band positive with slight population differences in staining intensity. Based on the C-banding results, four common banded chromosomal patterns were recognized in the populations of the Iranian house mouse. First, the largest CHRs was found in Zahedan and Zabol populations in all autosomes. Second, the smallest CHRs was observed in the chromosomes of subspecies M. musculus musculus from northeastern Iran. Third, in the specimens of subspecies Mus musculus castaneus, patterns of CHRs among the chromosomes was completely heterogeneous: some had large CHRs while others had very small CHRs. Fourth, small CHRs was observed in all chromosomes of subspecies Mus musculus domesticus from western and southern Iran, with the exception of two or three chromosomes, which had large CHRs.
1. Introduction
Chromosomal evolution is thought to occur through a random process of breakage and rearrangements, such as Robertsonian translocations, resulting in karyotype differences and change of gene organization (Bailey et al. Citation2004; Piálek et al. Citation2005). Chromosomal evolution in the house mouse, Mus musculus, has been extensively studied (Britton-Davidian et al. Citation2000; Piálek et al. Citation2005; Ezaz et al. Citation2006). This genus encompasses at least 40 species divided into four subgenera (Britton-Davidian et al. Citation2000). One species of the Mus subgenera is the house mouse, Mus musculus, which has four subspecies in Iran including M. m. domesticus, M. m. musculus, M. m. castaneus and M. m. bactrianus. These subspecies have the same standard karyotype with 20 pairs of acrocentric chromosomes (Silver Citation2001). Despite the use of modern molecular techniques, the presence and validity of the Iranian house mouse subspecies, e.g. M. m. castaneus, are still unresolved (Vanlerberghe et al. Citation1986; Orth et al. Citation1996; Darvish, Orth et al. Citation2006; Rajabi-Maham Citation2007). Additionally, molecular techniques are usually expensive, and may provide uncertain subspecies identification. Therefore, the cytogenetic method may be suitable for resolving taxonomic issues. Unlike the high morphological similarity in Murid species, the chromosomal characters of house mouse showed higher variability and were used to identify subspecies (Sharma et al. Citation2003; Romanenko et al. Citation2007).
Centromeric heterochromatin is most useful to identify populations and subspecies of house mouse (Rudra and Bhadur Citation2013). The centromeric domain of most mammalian chromosomes contains repeated DNA sequences that show variation among populations (Britten and Kohne Citation1968). In M. musculus, the major repetitive DNA component is the satellite DNA sequence family, which was first isolated by Kit (Citation1961). The major satellite is present on all M. musculus chromosomes with the exception of the Y chromosome, and constitutes around 10% of the M. musculus genome (Pardue and Gall Citation1969; Jones Citation1970). Sharma et al. (Citation2003) identified three types of M. terricolor in India by measuring centromeric heterochromatin. Cazaux et al. (Citation2014) showed three satellite DNA sequences within M. musculus – minor satellite, major satellite and telomeric location of chromosome (TLC) satellite – using FISH (fluorescence in situ hybridization); these appear to be physically very close to the primary constriction (i.e. the centromere) in these chromosomes. In comparison, the major satellite DNA sequences are localized to the pre-centromeric heterochromatin in M. musculus, and occupy a separate domain from minor satellite DNA sequences without any overlapping (Joseph et al. Citation1989). The latter study indicated high variability in the major satellite in populations of the house mouse. In the present study, we aimed to evaluate the centromeric region variations at the population of house mouse of Iran, to accurately determine the number of subspecies and their validity in the populations of the Iranian house mouse.
2. Materials and methods
2.1. Sampling data
A total of 103 individuals of M. musculus were captured from 20 localities in Iran. The sampling localities of house mouse are given in Figure and Table . The mice were caught with live-traps in farm buildings. Four morphometric characters including the length of body, tail, ear and hind foot were measured. The three subspecies were identified according to morphologic and morphometric characters from previous studies as indicated in Table (Darvish, Orth et al. Citation2006; Bonhomme et al. Citation2007; Shabani et al. Citation2010; Siahsarvie et al. Citation2012; Rajabi-Maham et al. Citation2012). All collected specimens are deposited in the Zoological Museum of Ferdowsi University of Mashhad (FUMZM).
Figure 1. Map of collection sites of M. musculus in Iran. Black boxes are marked with the name of the cities where mice were captured in this study.
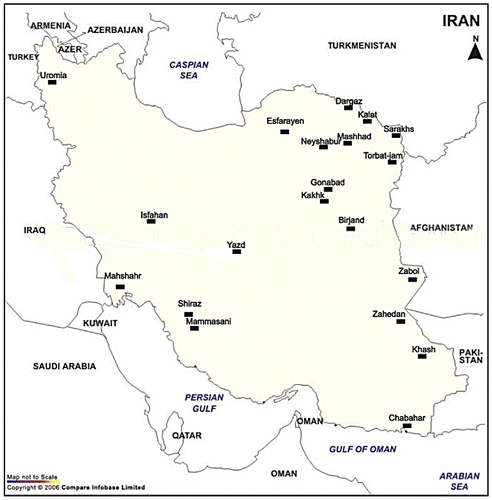
Table 1. Locality and taxon name of studied individuals of M. musculus.
2.2. Laboratory data
Mitotic chromosomes were prepared from bone marrow. The house mice were injected with vinblastine sulfate with hypotonic treatment following Yosida (Citation1973). The C-banding method was performed according to the BSG (barium/saline/Giemsa) method of Summer (Citation1972) with slight modifications. Ten to 30-day-old karyotype slides were treated with 0.2 N hydrochloric acid for 1 h at room temperature followed by 1–2 rinses in distilled water. The slides were treated in freshly prepared 5% aqueous solution of barium hydroxide (Ba(OH),-8H2O) at 60°C for 15 min, followed by thorough rinsing in distilled water. Slides were dried and incubated for 1 h at 60°C in 2 × SSC(1 × SSC: 0.15 M sodium chloride plus 0.015 M Tri-sodium citrate), pH 7.2. SSC treated slides were rinsed in distilled water and stained in 5% Giemsa (Merck, Germany) in phosphate buffer (pH 6.8) for 20 min.
About 50 to 100 metaphase spreads from each specimen were examined and at least 30 good chromosomal spreads were photographed using a 100× zoom digital CCD camera. The karyological characteristics of all specimens were prepared by karyological analysis software version 2 (Yu et al Citation2015). The karyological analysis software is available at http://mnh.scu.edu.cn/soft/blog/KaryoType (Sichuan University, China). The chromosomes were classified according to Levan et al. (Citation1964), The chromosomes were numbered on the basis of euchromatic long arms as per the recommendations of the Committee on Standardized Genetic Nomenclature of Mice (1972).The size of centromeric regions was measured by karyotype analysis software. To evaluate significance of differences among chromosomal of different populations we performed univariate analyses of variance (ANOVA) on chromosomal characters using SPSS 16.0 (www.ibm.com/software/analytics/spss).
3. Results
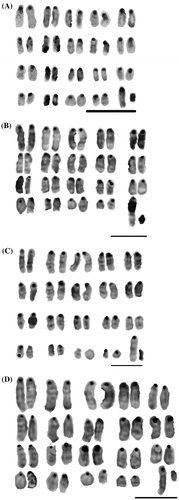
All the individuals of M. musculus demonstrated a diploid number of 2n = 40 including 38 acrocentric autosomes, a large acrocentric X and a small and dark acrocentric Y chromosome. In all the studied centromeric heterochromatin region sizes (CHRs) of the Iranian house mouse, Robertsonian translocation has not been observed. Due to similarity in size of centromeric heterochromatin on all chromosomes, Mashhad samples were used as standard for comparison between all samples. The results showed that the sizes of the centromeric region were variable. According to Table , chromosomes 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16, and 19 had the highest variation and chromosomes 1, 2, 3, 13, 17, 18, and X had the lowest variation in CHRs between specimens from different localities. In addition, the results indicated that chromosome 4 had the largest CHRs and chromosomes 14, 15, 16 and 19 showed the smallest CHRs in all studied specimens.
Table 2. Descriptive statistics of all chromosomes of all individual of house mouse for CHR regions.
C-banding patterns revealed extensive heterochromatin variations within and between populations of house mouse. Based on the C-banding method, four obvious banded chromosomal patterns were recognized in the Iranian house mouse. First, all chromosomes of Zahedan and Zabol populations showed larger CHRs than the other populations and in homologous chromosomes as well. Second, chromosomes of northeastern Iran populations, including eight locations, showed small CHRs in their chromosomes and patterns of C-positive heterochromatin presented similar type in all autosomal chromosomes. Third, chromosomal patterns of individuals of Mus musculus castaneus presented high variations in CHRs. Finally, CHRs for subspecies Mus musculus domesticus were small, with the exception of two or three chromosomes (usually chromosome 4, 10, and 13) and homologous chromosomes showed similar size of the C-banding region (Figure ). According to the ANOVA, all chromosomal characters were significantly different among different populations of house mouse (see more details in Appendix). In the C-band of all studied specimens one distinct point was recognized. The short arm of X chromosomes in all populations was invariably C-band positive. The telomere of long arms did not show prominent C-band positive staining in all populations The entire Y chromosome was consistently C-band positive in all populations.
4. Discussion
Based on our data, the Iranian house mouse karyotypes did not show any Robertsonian translocations and revealed that all house mouse chromosomes belong to chromosome type III. Cytogenetic studies in some species of Mus revealed that three types of karyotypes in this genus, designated as chromosome type I, II and III. In type III, all chromosomes are acrocentric with C-band positive minute perceptible short arms (Rudra and Bahadur Citation2013).
Previous studies showed a C-band variant on chromosome 1 whereas our results showed high variation in CHRs in chromosomes 4, 5, 6, 7, 8, 9, 10, 11, 12, 14, 15, 16, and 19 and low variation in CHRs in chromosomes 1, 2, 3, 13, 17, 18, and X between individuals of Iranian house mouse from different localities (Dev et al. Citation1973; Davidson Citation1989). According to Rudra and Bahadur (Citation2013), chromosome 19 of the Indian house mouse, Mus terricolor, represented large CHRs. Our results indicated that chromosomes 1, 4, 5, 6, 7, and 12 had large CHRs and the smallest CHRs were observed in chromosomes 15, 17, and 19 in all individuals of different localities. Additionally, C-band polymorphism of X chromosomes has been reported in of Mus (Sharma et al. Citation2003; Rudra and Bahadur Citation2013). This study showed that entire Y chromosomes of all individuals of house mouse were heterochromatic, and confirmed the previous study of (Rudra and Bahadur (Citation2013). The main object of this research was to find a simple and inexpensive taxonomic marker among chromosomal characters for recognizing subspecies of the Iranian house mouse that has been shown any unambiguous diagnostic morphological traits. Previously, the primary and secondary constrictions have been used for comparison between populations in mice (Dev et al. Citation1971; Eicher Citation1971; Forejt Citation1973; Vig and Richards Citation1992). Further studies showed that chromosomal characters are very useful for recognizing species in Rodentia (Malcon et al. Citation2007; Gunduz Citation2010). One of the chromosomal characters is size of the C-band region, which comprises three satellites including minor, major and TLC satellites (Cazaux et al. Citation2014). However, major satellites showed high variability in the house mouse (Southerne Citation1970; Cazaux et al. Citation2014; Rudra and Bahadur Citation2013). Constitutive heterochromatin has been shown to be highly polymorphic within and between genera of Mus (Piálek et al. Citation2005; Nadjafova Citation2008; Mitsainas et al. Citation2009; Karamysheva et al. Citation2010; Rubtsov et al. Citation2011). Our results showed that different C-banding patterns in each chromosome are characteristic for each subspecies of Mus musculus. The results showed small CHRs in all chromosomes and one common C-banding pattern in northeastern Iranian house mouse populations that were previously recognized as M. m. musculus (Darvish, Mirshamsi et al. Citation2006; Darvish, Orth, et al. Citation2006; Siahsarvie et al. Citation2012, Rajabi-Maham et al. Citation2012). M. musculus has at least three main subspecies (Boursot et al. Citation1993; Prager et al. Citation1998; Guénet and Bonhomme Citation2003; Geraldes et al. Citation2008; Bonhomme et al. Citation2011; Yonekawa et al. Citation2012). The standard karyotype of this species has 40 acrocentric chromosomes whereas some subspecies have lower diploid numbers as a result of Robertsonian whole-arm translocations (Gropp and Winking. 1981; Nuri et al. Citation2006; Rudra and Bahadur Citation2013).
Our results showed two C-banding patterns of M. m. castaneus: the first pattern in Zabol and Zahedan regions; and another pattern in populations from other localities including Khash, Esfahan, Shiraz and Yazd. Previous authors only mentioned that high variability occurs in populations of M. m. castaneus (Rajabi-Maham et al. Citation2012). This study showed similarity between the specimens of M. m. castaneus and M. m. bactrianus from Afghanistan. The type locality of M. m. bactrianus is Kandahar (Afghanistan); this subspecies has been reported from Iran using molecular study on nuclear genes (Din et al. Citation1996). The systematic status of the intermediate populations from eastern Iran, Afghanistan, Pakistan and northern India remains unclear (Darvish, Orth et al. Citation2006). In this study, remarkable banded chromosome of Zabol and Zahedan with the other populations of subspecies M. m. castaneus suggesting he specimen, from Zabol and Zahedan, probably is belonging to subspecies M. m. bactrianus. As has been mentioned, fourth banded chromosome pattern has been showed small CHRs except for a few chromosomes from western and southern Iran. According to Siahsarvie et al. (Citation2012) and Rajabi-Maham et al. (Citation2012), compared with the other subspecies of M. musculus, the population boundaries of M. m. domesticus are completely distinct using molecular studies. Sampling and supplementary studies are now needed to resolve ambiguous issues of the Iranian house mouse.
Funding information
This study was supported by a grant [no. 27137] from Ferdowsi University of Mashhad.
Disclosure statement
No potential conflict of interest was reported by the authors.
Acknowledgements
We are grateful to Dr M. Malekzadeh and Dr J. Davidian-Britton for their generous help and M. Mohammadyari, M. Mahmoodi, M. Habbiby, A. Khajea and A. Khosravi for their efforts in collecting the specimens.
References
- Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE. 2004. Hotspots of mammalian chromosomal evolution. Genome Biol. 5(4):R23.
- Bonhomme F, Orth A, Cucchi T, Rajabi-Maham H, Catalan J, Boursot P, Auffray JC, Britton-Davidian J. 2011. Genetic differentiation of the house mouse around the Mediterranean basin: matrilineal footprints of early and late colonization. P Roy Soc B-Biol Sci. 278(1709):1034–1043.
- Bonhomme F, Rivals A, Orth A, Grant G, Jeffreys AJ, Rj-Bois P. 2007. Species-wide distribution of highly polymorphic minisatellite markers suggests past and present genetic exchanges among house mouse subspecies. Genome Biol. 8(5):567–612.
- Boursot P, Auffray JC, Britton-Davidian J, Bonhomme F. 1993. The evolution of house mice. Annu Rev Ecol Syst. 24(97):119–152.
- Britten RJ, Kohne DE. 1968. Repeated DNA sequences in DNA. Science. 161(3841):529–540.
- Britton-Davidian J, Catalan J, Ramalhinho M, Ganem G, Auffray JC, Capela R, Biscoito M, Searle J, Mathias ML. 2000. Rapid chromosomal evolution in island mice. Nature. 403(6766):158–163.
- Cazaux B, Catalan J, Claude J, Britton-Davidian J. 2014. Non-random occurrence of robertsonian translocations in the house mouse (Mus musculus domesticus): Is it related to quantitative variation in the minor satellite. Cytogenet Genome. 8:28–31.
- Darvish J, Mirshamsi O, Kayvanfar N, Shakib NH. 2006. Diversity of the rodents of northeastern Iran. Iran J Anim Biosyst. 2(1):57–76.
- Darvish J, Orth A, Bonhomme F. 2006. Genetic transition in the house mouse Mus musculus of eastern Iranian plateau. Folia Zool. 55(4):349–357.
- Darvish J. 2008. Biosystematic approach to geographic variations of house mouse group, Mus musculus L.1766 Iranian. J Anim Biosyst. 4(1):31–54.
- Davidson MT. 1989. Centromeric chromosome variants. In: Lyon MF, Searle AG, editors. Genetic variant and strains of the laboratory mouse. 2nd ed. New York (NY): Oxford University Press. 417 p.
- Dev VG, Grewal MS, Miller DA, Kouri RE, Hutton JJ, Miller OJ. 1971. The quinacrine fluorescence karyotype of Mus musculus and demonstration of strain differences in secondary constrictions. Cytogenetics. 10(10):436–451.
- Dev VG, Miller DA, Miller OJ. 1973. Chromosome markers in Mus musculus strain differences in C-banding. Genetics. 75(4):663–670.
- Din W, Anand R, Boursot P, Darviche D, Dod B, Jouvin-Marche E, Orth A, Talwar G, Cazenave PA, Bonhomme F. 1996. Origin and radiation of the house mouse: clues from nuclear genes. J Evol Biol. 9(5):519–539.
- Eicher EM. 1971. The identification of the chromosome bearing linkage group XI1 in the mouse. Genetics. 69(2):267–271.
- Ezaz T, Stiglec R, Veyrunes F, Graves JAM. 2006. Relationships between vertebrate ZW and XY sex chromosome systems. Curr Biol. 16(17):736–743.
- Forejt F. 1973. Centromeric heterochromatin polymorphism in the house mouse. Chromosoma. 43:187–201.
- Geraldes A, Basset P, Gibson B, Smith KL, Harr B, Yu HT, Bulatova N, Ziv Y, Nachran MW. 2008. Inferring the history of speciation in house mice from autosomal, X-linked. Y-linked and mitochondrial genes. Mol Ecol. 17(24):5349–5363.
- Guenet JL, Bonhomme F. 2003. Wild mice an ever increasing contribution to a popular mammalian model. Trends Genet. 19(1):5467–5472.
- Gunduz I. 2010. Staggered chromosomal hybrid zones in the house mouse: relevance to reticulate evolution and speciation. Genes. 1(2):193–209.
- Jones K. 1970. Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature. 225(5236):912–915.
- Joseph A, Mitchell AR, Miller OJ. 1989. The organisation of the mouse satellite DNA at centromeres. Exp Cell Res. 183(2):494–500.
- Karamysheva TV, Bogdanov AS, Kartavtseva IV, Likhoshvay TV, Bochkarev MN, Kolcheva NE, Marochkina VV, Rubtsov NB. 2010. Comparative FISH analysis of c-positive blocks of centromeric chromosomal regions of Pygmy Wood Mice Sylvaemus uralensi (Rodentia, Muridae). Annu Rev Cell Dev Biol. 46(6):712–724.
- Kit S. 1961. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 3(3):711–716.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Malcon A, Smith F, Trifonor V. 2007. Mammalian karyotype evolution. Nature. 63(8):950–961.
- Mitsainas GP, Rovatsos MT, Giagia-Athanasopoulou EB. 2009. The HSR and heterochromatin distribution in natural populations of Mus musculus domesticus (Rodentia: Muridae) from Greece. Caryologia. 62(1):53–61.
- Nadjafova RS. 2008. Cytogenetic recognition of the common wood mouse, Sylvaemus sylvaticus s.1. (Mammals: Rodentia: Muridae) in European Russia. Compar Cytogenet. 2(1):1–6.
- Nuri Y, Moradi Gharakhaloo M, Olak E, Zkurt Þ, Bulut Þ, Kankili T, Olak R. 2006. The karyotypes of some rodent species (Mammalia: Rodentia) from Eastern Turkey and Northern Iran with a new record, Microtus schidlovskii Argyropulo, 1933, from Eastern Turkey. Turk J Zool. 30(2):459–464.
- Orth A, Elena L, Andrei K, Stephan B, Pierre B, Vo Nocolai, Francois B. 1996. L'espece polytypique Mus musculus en transcaucasie. Polytypic species Mus musculus in Transcucasia. CR Acade Sci paris. Sci de la vie/Lif Sci. 319:435–441.
- Pardue ML, Gall JG. 1969. Molecular hybridisation of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci USA. 63(2):600–604.
- Piálek J, Hauffe HC, Searle JB. 2005. Chromosomal variation in the house mouse: a review. Biol J Linn Soc. 84(2):535–563.
- Prager EM, Orrego C, Sage RD. 1998. Genetic variation and phylogeography of central Asian and other house mice, including a major new mitochondrial lineage in Yemen. Genetics. 150(2):835–861.
- Rajabi-Maham MH. 2007. Phylogéographie des souris du complex Mus musculus en Iran: Coalescence mitochondriale et diversite du chromosome Y [these]. [France]: Universite Montpellier II. p. 48.
- Rajabi-Maham H, Orth A, Siahsarvie R, Boursot P, Darvish J, Bonhomme F. 2012. The south-eastern house mouse Mus musculus castaneus (Rodentia: Muridea) is a polytypic subspecies. Biol J Linn Soc. 107(2):295–306.
- Romanenko SA, Volobouev VT, Perelman PL, Lebedev VS, Serdukova NA, Trifonov VA, Biltueva LS, Nie V, Obrien PCM, Bulatova NS, et al. 2007. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroidea, Rodentia) inferred from chromosomal painting and banding comparison. Chromosome Res. 15(2):283–297.
- Rubtsov NB, Karamysheva TV, Bogdanov AS, Likhoshvay TV, Kartavtseva IV. 2011. Comperative FISH analysis of C-positive regions of chromosomes of Wood Mice (Rodentia:Muridae). Russ J Genet. 47(9):1096–1110.
- Rudra M, Bahadur M. 2013. Heterochromatin variation among the populations of Mus terricolor Blyth, 1851 (Rodentia: Muridae) chromosome type I. Compar Cytogenet. 7(2):139–151.
- Shabani M, Darvish J, Mashkour M, Ghasemzadeh F, Mirshamsi O. 2010. Contemporary and sub-fossil house mice (Mus musculus Linnaeus, 1758) (Rodentia: Muridae) from Iran. Iran J Anim Biosyst. 6(2):45–54.
- Sharma T, Bardhan A, Bardhan M. 2003. Reduced meiotic fitness in hybrids with heterozygosity for heterochromatin in the speciating Mus terricolor complex. Bioscience. 28(2):137–143.
- Siahsarvie R, Auffray JC, Darvish J, Rajabi-Maham H, Yu HT, Agret S, Bonhomme F, Claude J. 2012. Patterns of morphological evolution in the mandible of the house mouse Mus musculus (Rodentia: Muridae). Biol J Linn Soc. 105:635–647.
- Silver LM. 2001. Chromosomes: mouse genetics, concepts and applications. Chapter 5 [revised 2004 Aug; 2008 Jan]. Oxford: Oxford University Press. 357 p.
- Southerne M. 1970. Base sequence and evolution of guinea pig a-satellite DNA. Nature. 227(5236):794–798.
- Summer AT. 1972. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 75:304–306.
- Vanlerberghe F, Dod B, Boursot P, Bellis M, Bonhomme F. 1986. Absence of Y chromosome introgression across the hybrid zone between Mus musculus domesticus and Mus musculus musculus. Genet Res. 48(3):91–197.
- Vig BK, Richards BT. 1992. Formation of primary constriction and heterochromatin in mouse does not require minor satellite DNA. Cell Res. 201(2):292–298.
- Yonekawa H, Sato JJ, Suzuki H, Moriwaki K. 2012. Origin and genetic status of Mus musculus molossinus: a typical example for reticulate evolution in the genus Mus. Isolation and gene flow: inferring the speciation history of European house mice. Mol Ecol. 20(2):5248–5264.
- Yosida TH. 1973. Frequencies of chromosome polymorphism (pairs no. 1, 9 and 13) in the black rat, Rattus rattus diardii (Rodentia, Muridae). Chromosoma. 36:256–262.
- Yu Y, Altınordu F, Peruzzi L, He XJ. 2015. KaryoType 2.0. Sichuan University, China. Available at: http://mnh.scu.edu.cn/soft/blog/KaryoType