Abstract
The present study reports the chromosome number and meiotic behavior of 11 populations belonging to four species of Phlomoides (L.) Moench. All populations showed the diploid chromosome number 2n = 2x = 22; consistent with the proposed base number of x = 11. Almost all taxa studied here displayed regular bivalent pairing and chromosome segregation at meiosis. However, meiotic abnormalities observed included varied degree of fragmented chromosomes, laggards and bridge in anaphase I to telophase II, precocious division of centromeres, asynchronous nucleus and cytomixis. In addition, a survey on chromosome counts of different species belonging to the genus Phlomoides throughout the world is presented and the base chromosome numbers and polyploidy levels discussed here.
Introduction
The plant family Lamiaceae Martinov (=Labiatae Adans., the mint family) has a worldwide distribution and comprises more than 7200 species across approximately 233–263 genera, of which subfam. Nepetoideae contains over 50% of all species (Harley et al. Citation2004). Phlomideae Mathiesen contains 278 species, whereas Kamelin and Makhmedov (Citation1990) recognized about 250 species within the component genera. This tribe, originally suggested to comprise six genera (Scheen et al. Citation2010; Mathiesen et al. Citation2011), is a complex group within Lamioideae: Eremostachys Bunge, Lamiophlomis Kudô, Notochaete Benth., Phlomis L., Phlomoides (L.) Moench, and Pseuderemostachys Popov. The species are distributed from Europe to Mongolia, China, and India with the highest number of species found in Central Asia, Afghanistan and Iran (Irano-Turanian and Himalayan regions). They comprise elements of subalpine and alpine vegetation with some species growing in desert conditions. Phlomis L. is a large group of lamioid labiates. As early as 1794, Moench recognized morphological differences within Phlomis that he believed to be characteristic enough to split the taxon into two separate genera, Phlomis and Phlomoides Moench (Moench Citation1794). These morphological differences have also been recognized by many other authors (Link Citation1829; Bentham Citation1832–1836; Boissier Citation1879; Briquet Citation1895–1897; Kamelin and Makhmedov Citation1990; Mathiesen et al. Citation2011). The position of Eremostachys within tribe Phlomideae has also been assessed in recent molecular phylogenetic studies of the subfamily Lamioideae (Scheen et al. Citation2010; Bendiksby et al. Citation2011). Molecular data show the tribe Phlomideae as an assembly of closely related genera, confirming previous assumptions (Scheen et al. Citation2010; Bendiksby et al. Citation2011; Salmaki et al. Citation2012a, 2012b). Molecular phylogenetic studies by Salmaki et al. (Citation2012b) provided definite evidence on the close relationship between Eremostachys and Phlomoides based on the sequence of nuclear ribosomal DNA (ITS) and cpDNA. One of the main reasons for inclusion of Eremostachys in Phlomoides was the large number of morphologically transitional species between Phlomoides s.str. and the Eremostachys core group (Salmaki et al. Citation2012a). In its new definition, Phlomoides contains about 150–170 species (Kamelin and Makhmedov Citation1990) and its distribution area extends from Central Europe to the Russian Far East. The major centers of diversity of Phlomoides are Central Asia, China and the Flora Iranica area (Afghanistan, Iran, W Pakistan, SW Turkmenistan, NE Iraq).
Most of the previous cytological studies in the genus Phlomoides and related genera have concentrated on the chromosome count (Zhukova Citation1967; Kartashova et al. Citation1974b; Aryavand Citation1975; Raina and Ashruf Citation1981; Saggoo and Bir Citation1982, Citation1986; Saggoo Citation1983; Astanova Citation1984; Jee et al. Citation1985, 1989; Zakirova and Nafanailova Citation1988; Ma et al. Citation1990; Probatova Citation2006), with little work focused on detailed karyological criteria for taxonomic purposes. Studies on the impact of karyotypic and meiotic behavior data on the interspecific and phylogenetic relationships in the genus are still limited. To the best of our knowledge, meiotic behavior is almost absent in the Phlomoides. Even basic cytogenetic studies in this genus are few, and only approximately 17% of the species have had their chromosome numbers reported. Increasing information on the meiotic behavior of Phlomoides species may give important insights on the numerical and structural chromosome changes involved in the evolution of the genus. Since knowledge of the cytology is essential for understanding interspecific relationships, the aim of this research was to investigate the chromosome number and meiotic behavior of four species of Phlomoides (P. azerbaijanica (Rech.f.) Kamelin & Makhm., P. laciniata (L.) Kamelin & Makhm., P. laevigata (Bunge) Kamelin & Makhm. and P. macrophylla (Benth.) Kamelin & Makhm.) which had not previously been analyzed and to try to increase the basic cytogenetic knowledge of the genus.
This article follows previous cytological studies in Iran (Ranjbar et al. Citation2010; Sheidai et al. Citation2010; Ranjbar et al. Citation2011; Ranjbar and Mahmoudi Citation2013a, Citation2013b).
Materials and methods
Description of database
Chromosome counts were collected from a several online databases: Index to Plant Chromosome Numbers (IPCN; Goldblatt and Johnson Citation1979) and the global Chromosome Counts Data Base (CCDB; Rice et al. Citation2015) and an array of miscellaneous sources such as floras and other scientific manuscripts. Some chromosome records based on online databases and literature have been presented in Table . Each record in the database includes the following data: the name of the taxon as published in the original source; mitotic or meiotic chromosome number; ploidy level; and the name of the person who counted chromosomes.
Table 1. List of previous chromosome counts reported for the genus Phlomoides.
Plant material
Plant material collection was done in several different regions of Iran. Young flower buds were obtained in 11 populations of four Phlomoides species growing wild in this region, namely Phlomoides azerbaijanica, P. laciniata, P. laevigata and P. macrophylla. The voucher specimens are deposited in Herbarium of Bu-Ali Sina University (BASU), details of which are given in Table .
Table 2. Collection data of 11 populations of Phlomoides.
Cytological studies
Randomly selected plants in the ideal stage for meiotic studies were collected and fixed in 96% ethanol, chloroform and propionic acid (6:3:2) for 24 h at room temperature, and then washed and preserved in 70% ethanol at 4°C until used. Microsporocytes were prepared by squashing and stained with 2% acetocarmine. Chromosome numbers were determined in five individuals of each population during diakinesis. The meiotic chromosome association was evaluated in at least 20 diakinesis cells. A minimum of 100 metaphase/diakinesis pollen mother cells (PMCs) and 500 anaphase/telophase cells were analyzed for data collection. Meiotic stages were photographed by a BX-51 Olympus microscope equipped with a 3030 digital camera.
Phenetic studies
11 populations of different species of Phlomoides from several different regions of Iran were used as operational taxonomic units (OTUs) (Table ). A numerical taxonomic analysis of the different individuals from these populations was carried out based on 20 quantitative cytogenetic characters (Table ). Data were entered into Microsoft Excel 2007 (Microsoft, Redmond, WA, USA). This spreadsheet was later converted into a file format suitable for phenetic analysis by MVSP software version 3.2 (CitationKovach 1985–2002). Principal coordinate analysis (PCoA) was carried out using MVSP, with a matrix of standardized data. The data were standardized to eliminate the distorting effects in the output results caused by different measurement scales. Standardization was performed by subtracting the character mean and dividing by the standard deviation. For PCoA, an average-distance-matrix of standardized data was obtained. The average distance was used because the dataset contained both metric and binary (mixed) data. The distance matrix was double centered and the eigenvectors were calculated and plotted. The PCoA gives the distances between OTUs rather than the correlation between the characters.
Table 3. Number of pollen mother cells (PMCs) analyzed and percentage of PMC meiotic behavior in Phlomoides.
Results and discussion
Meiotic behavior
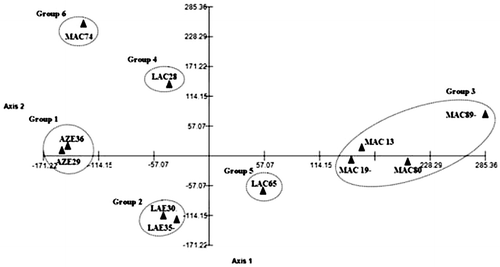
Meiotic behavior was determined in 55 individuals belonging to 11 populations of four species. A summary of their cytological features is given in Table , and the chromosomes are illustrated in Figures 1–51. A total of 3905 diakinesis/metaphase I (D/MI), 2065 anaphase I/telophase I (AI/TI), 1426 metaphase II (MII) and 2729 anaphase II/telophase II (AII/TII) cells were analyzed. Almost all taxa studied here displayed regular bivalent pairing and chromosome segregation at meiosis. However, meiotic irregularities observed in different populations included fragmented chromosomes, laggards and bridge in anaphase I to telophase II, precocious division of centromeres, asynchronous nucleus and cytomixis, which have been discussed below. All data were obtained from meiotic behavior analyzed by MVSP version 3.2. They showed interspecific and intraspecific variation that resulted in six main groups (Figure ). Group 1 included AZE36 and AZE29 populations, group 2 included LAE35, LAE30 populations and group 3 included MAC13, MAC19, MAC89 and MAC80 populations. LAC28, LAC65 and MAC74 populations individually formed groups 4, 5, 6, respectively. It seems that among the cytogenetic characters studied (Table ), a various degrees of abnormality, for example high score in the formation of cytomixy, precocious segregation, laggard chromosome and bridge, played decisive roles in differentiating populations. Group 1 with two populations is cytologically related to P. azerbaijanica, group 2 with two populations is related to P. laevigata and group 3 with four populations is related to P. macrophylla. Group 4, which comprises LAC28 population, is separated from group 5 by its higher frequency of precocious segregation, laggard chromosomes and bridge. The only population in group 6, MAC74, is separated from the group 3 by its low frequency of cytomixy in D/MI and AI/TI.
Results from PCoA analysis represented variation between different populations of each Phlomoides species. MAC74 in particular is quite isolated from the other MAC samples. However, these differences were not strong enough to justify a separation at interspecific level leading to a new species or even at intraspecific level for separating a new subspecies or a new variety. Constant genomic changes through specific gene interaction (Kumar and Bennetzen Citation1999; Bennetzen Citation2002) and the synthesis of new cellular components (Kimura et al. Citation1999) such as required for the new adaptive complexes is essential for species survival in a constantly changing environment (Cai and Xu Citation2007). Variability of characters is a consequence of several cytogenetic interactions that regulate gene expression pattern (Liu and Wendel Citation2003; Levy and Feldman Citation2004). The significance of meiosis in providing the platform for synthesis of morphological features is essential for species adaptation in new environments. Meiotic behavior helps to assess species potentials for reproductive success, genetic variability, biodiversity and survival in new environments (Boff and Schifini-Wittmann Citation2002).
Different species at the population level indicate the occurrence of different frequencies of meiotic abnormalities in the form of cytomixis, unoriented bivalents, chromatin stickiness, chromatin bridges and laggards, which reflects intraspecific genetic diversity (Baptista-Giacomelli et al. Citation2000; Sheidai et al. Citation2003). Many causes may lead to the occurrence of meiotic irregularities, such as lack of chromosomal homology resulting from hybridization, polyploidy, genetic and environmental factors (Pagliarini Citation1990).
Laggard chromosomes and precocious migration of chromosomes to the poles
Spindle fibers, which arrange chromosomes on the equatorial plate in order to gather them as a group, emerge at mid-prometaphase in both meiosis I and meiosis II to be attached at specific sites, called kinetochores, at metaphase, and separate the chromosomes/chromatids at anaphase (Caetano-Pereira and Pagliarini Citation2001; Pawan Kumar et al. Citation2013). Normal bipolar spindles ensure regular chromosome segregation; four types of abnormal spindle may exist, resulting in the inability to form two daughter nuclei, affecting viable gamete production and balanced gamete constitution (Shamina et al. Citation2003; Pawan Kumar et al. Citation2013). Multipolar or polyarchal spindles, one type of spindle aberration, constitute an integrated system of some half-spindles (arches) which cause chromosomes to distribute randomly among these arches and move to their poles at anaphase (Shamina et al. Citation2003). Abnormal chromosome segregation, presence of micronuclei in microspores and formation of polyads resulting from multipolar meiocytes are manifestations of this irregularity (Caetano-Pereira and Pagliarini Citation2001; Mansuelli et al. Citation1995). In this study, the majority of cells showed normal spindle formation, which resulted in regular arrangement of bivalents at the spindle plate during metaphase I and II. However, the presence of laggards as a consequence of abnormal chromosome segregation at anaphase I and II has been also confirmed in the studied taxa, ranging from 1.3% in AZE29 to 32.63% in MAC13. Most populations of P. macrophylla (MAC13: 32.63%, MAC80: 11.44%, MAC74: 5.13%, MAC89: 7.53%) and P. laciniata (LAC65: 6.63%, LAC28: 2.13%), and only one population from each of P. azerbaijanica (AZE29: 1.3%) and P. laevigata (LAE30: 3.03%) showed this abnormality (Figures , and ). Another abnormality related to irregular chromosome segregation, according to Sayuri Utsunomiya et al. (Citation2005), is precocious migration of chromosomes to the poles in metaphase I and II, which was observed in all populations of P. macrophylla (MAC13: 10.13%, MAC80: 8.42%, MAC74: 2.13%, MAC89: 1.63%), P. laciniata (LAC65: 6.66%, LAC28: 6.33%), P. azerbaijanica (AZE36: 4.3%, AZE29: 2.3%) and P. laevigata (LAE30: 2.23%, LAE35: 9.29%) with different percentage of precocious migration to the poles per individual (Figures , , and ).
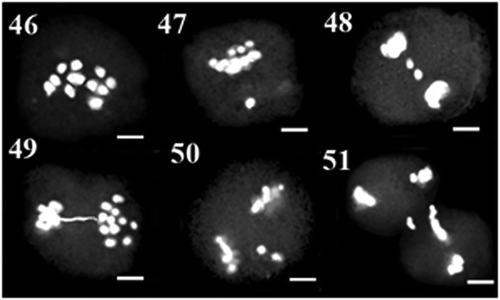
Figure 1–15. 1–6: Meiosis in MAC74 population: 1: diakinesis; 2: anaphase I with laggard chromosomes; 3: telophase I with fragmented chromosomes; 4: metaphase II with precocious chromosome; 5: anaphase II with bridge and precocious chromosome; 6: telophase II with bridge. 7–10: Meiosis in MAC80 population: 7: diakinesis; 8: metaphase I with precocious chromosome; 9: cytomixis in diakinesis; 10: anaphase I with laggard chromosomes. 11–15: Meiosis in MAC13 population: 11: diakinesis; 12: metaphase I with precocious chromosome; 13: anaphase I with bridge; 14: anaphase I; 15: cytomixis in metaphase I. Scale bars: 5 μm.
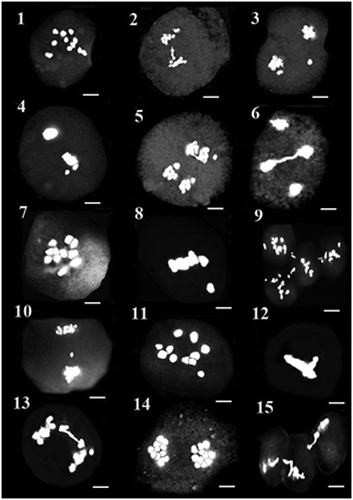
Figure 16–30. 16–21: Meiosis in MAC89 population: 16: diakinesis; 17: metaphase I with precocious chromosome; 18: cytomixis in metaphase I; 19: anaphase I with bridge and fragment chromosome; 20: cytomixis in metaphase II; 21: anaphase I with laggard. 22–25: Meiosis in LAE35 population: 22: metaphase I with precocious chromosome; 23: metaphase II with asynchronous nuclei; 24: cytomixis in diakinesis; 25: telophase I with bridge. 26–30: Meiosis in LAC28 population: 26: metaphase I with precocious chromosome; 27: anaphase I; 28: metaphase II with asynchronous nuclei and precocious chromosome; 29: anaphase II with bridge; 30: cytomixis. Scale bars: 5 μm.
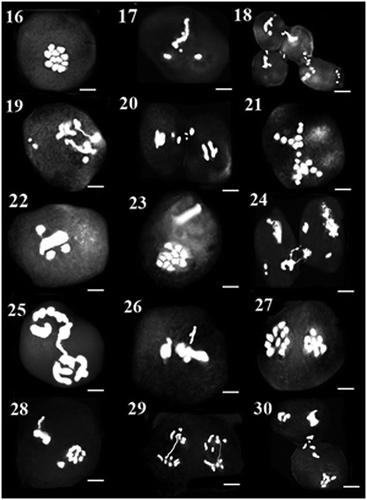
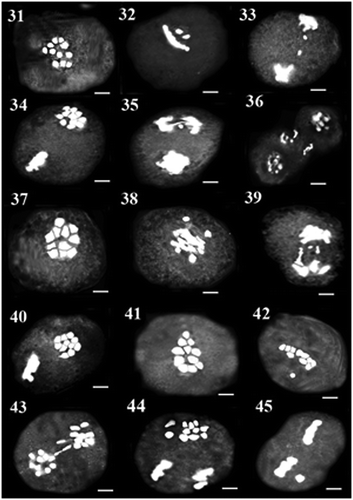
Figure 31–45. 31–33: Meiosis in AZE29 population: 31: diakinesis; 32: metaphase I with precocious chromosome; 33: anaphase I with laggard. 34–36: Meiosis in MAC13 population: 34: metaphase II with asynchronous nuclei; 35: anaphase II with laggard and bridge; 36: cytomixis in diakinesis. 37–40: Meiosis in LAC65 population: 37: diakinesis; 38: metaphase I with fragment chromosome; 39: anaphase I with laggard; 40: metaphase II with asynchronous nuclei. 41–45: Meiosis in LAE30 population: 41: diakinesis; 42: metaphase I with precocious chromosome; 43: anaphase I with laggard and bridge; 44: asynchronous nuclei; 45: metaphase II with precocious chromosome. Scale bars: 5 μm.
Cytomixis
The phenomenon of cytomixis consists in the migration of chromosomes between meiocytes through cytoplasmic connection. Since cytomixis creates variation in the chromosome number of the gametes, it could be considered a mechanism of evolutionary significance (Ghaffari Citation2006; Ranjbar et al. Citation2011). This phenomenon was observed in some populations of P. laciniata (LAC28: 1.03%), P. laevigata (LAE35: 0.89%) and P. macrophylla (MAC13: 3. 15%) in metaphase I, II, anaphase I and telophase II cells with different percentage of cytomixis per individual (Figures , , and ).
Chromosome stickiness and bridges
Phenotypic manifestation of chromosome stickiness, i.e. slight to intense clustering during any phase of the cell cycle, can be highly variable, and in severe cases pyknotic nuclei may be observed, resulting from the lack of chromosome separation, culminating in full chromatin degeneration (Mendes-Bonato et al. Citation2001). Both genetically controlled stickiness and stickiness resulting from environmental agents have been reported in many plant species (Baptista-Giacomelli et al. Citation2000). Among hypotheses which have been suggested to explain the phenomenon, Gaulden (Citation1987) postulated that stickiness results from mutation in structural genes or direct action of mutagens related to non-histone proteins (topoisomerase II and peripheral proteins), whose functions are necessary for separation and segregation of chromatids. In the populations analyzed, in diakinesis and metaphase I stages, several chromosomes to the entire chromosome complement have been involved in stickiness phenomena. MAC13 (1.03%) and AZE36 (41.2%) had the lowest and highest rates of stickiness, respectively. One (Figure ) to several (Figure ) bridges, resulting from chromatin stickiness, were recorded at both anaphases. Chromosome stickiness and bridges were observed in some populations of P. macrophylla (MAC13: 1.03%, MAC74: 12.02%, MAC89: 2.15%) and all populations of P. laciniata (LAC65: 2.69%, LAC28: 2.73%), P. azerbaijanica (AZE36: 41.2%, AZE29: 1.2%) and P. laevigata (LAE30: 3.43%, LAE35: 3.19%) with different percentage of chromosome stickiness and bridges per individual (Figures , , and ).
Chromosome data analysis
In the genus Phlomoides, our own counts for P. azerbaijanica, P. laciniata, P. laevigata and P. macrophylla are all 2n = 22 (Table ). The present determination of the chromosome numbers in P. azerbaijanica and P. laevigata are the first report. The observation of the present study as well as the available data on chromosome number in the literature for the genus Phlomoides (Table ) shows that, among the approximately 25 taxa with the chromosome number counts (approximately 17–19% of species with known chromosome numbers), the diploid taxa represent 89.6% of the whole, while the diploid and polyploid represent 10.34%. There are records for approximately 17–19% of species with known chromosome numbers.
Chromosome numbers in the genus Phlomoides as a whole show considerable variation (Table ). The available data on chromosome number in the genus confirmed that there are different chromosome numbers in genus Phlomoides (2n = 12, 14, 21, 20, 22, 24, 28, 42, 46 and 88) (Zhukova Citation1967; Kartashova et al. Citation1974a; Aryavand Citation1975; Raina and Ashruf Citation1981; Saggoo and Bir Citation1982, Citation1986; Saggoo Citation1983; Astanova Citation1984; Jee et al. Citation1985, 1989; Zakirova and Nafanailova Citation1988; Ma et al. Citation1990; Probatova Citation2006). The highest frequent chromosome number is 2n = 88, reported for P. macrophylla (Saggoo and Bir Citation1982, Citation1986; Saggoo Citation1983) from S Asia (India). These numbers probably represent aneuploid members that establish communities surrounding the diploid and polyploid species. The diploid number 2n = 22 is known in most of the taxa belonging to the genus. Therefore, x = 11 is the prevailing basic number in the genus. We report here the chromosome numbers of 11 populations of four species belong to genus Phlomoides, which were collected from different localities in Iran (Table ). Our results showed that all populations were diploid and possessed 2n = 2x = 22 chromosome number, consistent with the proposed base number of x = 11. On the basis of published counts for genus Phlomoides together with the reports presented here, we have verified that x = 11 is the most common number for this genus.
Chromosome cytology has a rich history in plant systematic and evolutionary biology. These data have been widely used to evaluate the evolutionary pattern of chromosome number change and to estimate the base chromosome number of clades of interest (Mayrose et al. Citation2014). Chromosome numbers have also been extensively utilized as an important phylogenetic character in the context of cytotaxonomy (Chatterjee and Kumar Sharma Citation1969; Schlarbaum and Tsuchiya Citation1984; Guerra Citation2012; Ranjbar and Mahmoudian Citation2015; Ranjbar et al. Citation2015). Phylogenetic relationships in genus Phlomoides have been intensively studied using molecular sequence data (Scheen et al. Citation2010; Bendiksby et al. Citation2011; Mathiesen et al. Citation2011; Salmaki et al. Citation2012b), providing a framework for assessing cytological patterns in the genus. Chromosome counts for the genus are scattered in the literature, many of them published under genus Phlomis and Eremostachys that are now relegated to synonymy. Azizian and Culter (Citation1982) reported that within Phlomis, two distinct groups can be recognized using chromosome data: Phlomis section Phlomis has 2n = 20 and Phlomis section Phlomoides has 2n = 22. Li-Qin et al. (Citation2007) also confirmed the basic chromosome number x = 11 for Phlomis sect. Phlomoides. Eremostachys also has chromosomes corresponding to those of Phlomis sect. Phlomoides. The chromosome numbers reported here for genus Phlomoides support the earlier report. This concurs renders further support a split of the genus Phlomis into two separate groups that are recognized as the genera Phlomoides and Phlomis (Mathiesen et al. Citation2011) and transfer of several species of Eremostachys to Phlomoides (Salmaki et al. Citation2012b). For genus Phlomoides then, we accept the hypothesis that x = 11 is the ancestral basic chromosome number.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by Bu Ali Sina University grant number [1164].
Acknowledgements
The authors would like to thank Dr. A. Papini for their generous comments and support during the review process. They would also like to thank the anonymous reviewers for their helpful and constructive comments that greatly contributed to improving the final version of the paper.
References
- Aryavand A. 1975. Contribution to the cytotaxonomic study of some angiosperms of Iran. Botaniska Notiser. 128(3):299–311.
- Astanova SB. 1984. Chromosome numbers in the species of the families Alliaceae, Asteraceae, Caryophyllaceae, Ebenaceae, Linaceae, Oleaceae, Lamiaceae from Tadjikistan. Bot. Žurnal SSSR. 69(11):1563–1564.
- Astanova SB, Kochkareva TF. 1982. Kariosistematicheskoe izuchenie vidov roda Eremostachys Bunge - pustynnokolosnik flory Tadzhikistana [A karyosystematic study of species Eremostachys Bunge - Flora of Tajikistan]. Izvestiia Akademii Nauk Tadzhikskoi SSR: Otdelenie Biologicheskikh Nauk. 2(87):3–8.
- Astanova SB, Kochkareva TF. 1985. Kariosistematicheskoe isuchenie endemichnykh dlja Pamiro-Alaja vidov roda Pustynnokolosnik (Eremostachys Bunge) A karyosystematic study of endemic species from Pamir-Alai (Eremostachys Bunge). Izvestiia Akademii Nauk Tadzhikskoi SSR: Otdelenie Biologicheskikh Nauk (Dushanbe). 1(94):65–68.
- Azizian D, Culter DF. 1982. Anatomical, cytological and phytochemical studies on Phlomis L. and Eremostachys Bunge (Labiatae). Bot. J. Linn. Soc. 85(4):249–281.10.1111/boj.1982.85.issue-4
- Baptista-Giacomelli FR, Pagliarini MS, Almeida JL. 2000. Meiotic behavior in several Brazilian oat cultivars (Avena sativa L.). Cytologia. 65(4):371–378.10.1508/cytologia.65.371
- Bendiksby M, Thorbek L, Scheen AC, Lindqvist C, Ryding O. 2011. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon. 60(2):471–484.
- Bennetzen JL. 2002. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 115(1):29–36.10.1023/A:1016015913350
- Bentham G. 1832–1836. Labiatarum genera et species. London: James Ridgway & Sons.
- Boff T, Schifini-Wittmann MT. 2002. Pollen fertility and meiotic behavior in accessions and species of Leucaena. Tropical Grasslands. 36(1):54–58.
- Boissier E.1879. Flora orientalis, vol. 4. Geneva: H. Georg.
- Briquet J. 1895–1897. Labiatae. Eremostachys Bunge, Phlomis L. In: Engler A, Prantl K,, editors. Die natürlichen Pflanzenfamilien. Leipzig: Engelmann. p. 246–249.
- Caetano-Pereira CM, Pagliarini MS. 2001. A new meiotic abnormalityin Zea mays: multiple spindles associated with abnormal cytokinesis in both divisions. Genome. 44(5):865–871.10.1139/g01-079
- Cai X, Xu SS. 2007. Meiosis-driven genome variation in plants. Curr Genomics. 8(3):151–161.10.2174/138920207780833847
- Chatterjee T, Kumar Sharma A. 1969. Cytotaxonomy of cichorieae. Genetica. 40(1):577–590.10.1007/BF01787382
- Chuksanova NA, KaplanbekovaSA. 1971. Chromosome numbers in certain species of Labiatae Juss and Scrophulariaceae Lindl. indigenous to the USSR (in Russian). Bot. Žurnal. 56: 522–528.
- Dhawan M, Gupta BK. 1983. In IOPB chromosome number reports LXXX. Taxon. 32(3):504.
- Dobea C, Hahn B, Morawetz W. 1997. Chromosomenzahlen zur Gefässpflanzen-Flora Österreichs. Linzer Biologische Beiträge. 29(1):5–43.
- Gaulden ME. 1987. Hypothesis: some mutagens directly alter specific chromosomal proteins (DNA topoisomerase II and peripheral proteins) to produce chromosome stickiness, which causes chromosome aberrations. Mutagenesis. 2(5):357–365.10.1093/mutage/2.5.357
- Ghaffari SM. 2006. Occurrence of diploid and polyploidy microspores in Sorgum bicolor (Poaceae) is the result of cytomixis. Afr. J. Biotechnol. 5(16):1450–1453.
- Gill LS. 1970. Cytological observations on West-Himalayan Labiatae: Tribe Stachydeae. Phyton (Buenos Aires). 27(2):177–184.
- Gill LS. 1972. Chromosome numbers in West-Himalayan bicarpellate species II. -. Bull. Torrey Bot. Club. 99(1):36–38.10.2307/2484241
- Gill LS. 1984. The incidence of polyploidy in the West-Himalayan Labiatae. Revue de Cytologie et de Biologie Végétales, le Botaniste. 7(1):5–16.
- Goldblatt P, Johnson DE, editors. 1979. Index to plant chromosome numbers (IPCN). Available from: http://www.tropicos.org/Project/IPCN.
- Guerra M. 2012. Cytotaxonomy: the end of childhood. Plant Biosyst. 146(3):703–710.
- Harley RM, Atkins S, Budantsev AL, Cantino PD, Conn BJ, Grayer R, Harley MM, de Kok R, Krestovskaya TV, Morales R. 2004. Labiatae. In: Kubitzki K, Kadereit JW, editors. The families and genera of vascular plants. Berlin, Heidelberg: Springer. p. 167–275.
- Jee V, Dhar U, Kachroo P. 1985. Chromosomal conspectus of some alpine-subalpine taxa of Kashmir Himalaya. Chromosome Inf. Serv. 39:33–35.
- Jee V, Dhar U, Kachroo P. 1987. Addition to the cytological conspectus of alpine–subalpine flora of Kashmir Himalaya. Chromosome Inf. Serv. 43:7–9.
- Jee V, Dhar U, Kachroo P. 1989. Cytogeography of some endemic taxa of Kashmir Himalaya. Proc. Nat. Acad. Sci. 55(3):177–184.
- Kamelin RV, Makhmedov AM. 1990. Sistema roda Phlomoides (Lamiaceae) [The system of the genus Phlomoides (Lamiaceae)]. Bot. Žurnal (Moscow & Leningrad). 75: 241–250.
- Kartashova NN, Malakhova LA, Kozlova AA. 1974a. Study of the chromosomes of the representatives of the Ob region flora. I. Number of chromosomes of the Tomsk district. Nauchnye Doklady Vysshei Shkoly Biol. Nauki. 4:114–119.
- Kartashova NN, Malakhova LA, Koslova AA, Dubrova NA. 1974b. Chisla chromosom u rjada polesnykh rastenij is prirodnykh populjacij flory Priob’ja. Biol. Biofisika TomskDostupné z. Available from: http://mobot.mobot.org/W3T/Search/ipcn.html.47–53.
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 1999. 13S condensing actively reconfigures DNA by introducing global positive writhe: Implications for chromosome condensation. Cell. 98(2):239–248.10.1016/S0092-8674(00)81018-1
- Kovach W. 1985-2002. Institute of Earth Studies, University College of Wales, Aberystwyth, (Shareware), MVSP Version 3.2, Kovach Computing Services.
- Krasnikov AA, Schaulo DN. 1990. Chromosome numbers in representatives of some families of vascular plants in the flora of the Novosibirsk region. II. Bot. Žurnal. (Moscow & Leningrad) 75(1): 118–120.
- Kumar A, Bennetzen JL. 1999. Plant retrotransposons. Ann. Rev. Genet. 33:479–532.10.1146/annurev.genet.33.1.479
- Lee YN. 1972. Chromosome number of flowering plants in Korea (4). J. Korean Res. Instit. Better Living. 8:41–51.
- Levy AA, Feldman M. 2004. Genetic and epigenetic reprogramming of the wheat genome upon allopolyploidization. Biol. J. Linn. Soc. 82:607–613.10.1111/j.1095-8312.2004.00346.x
- Link HF.1829. Handbuch zur Erkennung der nutzbarsten und am häufigsten vorkommenden Gewächse, Vol. 2. Berlin: S J Joseephy.
- Li-Qin F, Yue-Zhi P, Xun G. 2007. Anatomical, cytological and phytochemical studies on Phlomis L. and Eremostachys Bunge (Labiatae). Acta Phytotaxonomica Sinica. 45(5):627–632.
- Liu B, Wendel JF. 2003. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 29(3):365–379.10.1016/S1055-7903(03)00213-6
- Ma Xh, Xq Ma, Li N. 1990. Chromosome observation of some drug plants in Xinjiang. Acta Bot. Boreali-Occidentalia Sinica. 10(3):203–210.
- Mansuelli RW, Tanimoto EY, Brown C, Comai L. 1995. Irregular meiosis in a somatic hybrid between Solanum bulbocastanum and S. tuberosum detected by species-specific PCR markers and cytological analysis. Theor. Appl. Genet. 91(3):401–408.
- Mathiesen C, Scheen AC, Lindqvist C. 2011. Phylogeny and biogeography of the lamioid genus Phlomis (Lamiaceae). Kew Bull. 66(1):1–17.
- Mayrose IS, Zhan H, Rothfels CJ, Arrigo N, Barker MS, Rieseberg LH, Otto SP. 2014. Methods for studying polyploid diversification and the dead end hypothesis: a reply to Soltis. New Phytol. 206(1):27–35.
- Mendes-Bonato AB, Pagliarini MS, Silva N, Valle C. 2001. Meiotic instability in invader plants of signal grass Brachiaria decumbens Stapf (Gramineae). Acta Sci. 23(2):619–625.
- Moench C. 1794. Methodus plantas horti botanici et agri Marburgensis, a staminum situ describendi. Marburg: Officina Nova Libraria Academiae.
- Pagliarini MS. 1990. Meiotic behavior and pollen fertility in Aptenia Cordifolia (Aizoaceae). Caryologia. 43(2):157–162.10.1080/00087114.1990.10796994
- Pawan Kumar R, Kumar P, Singhal VK. 2013. Spindle irregularities, chromatin transfer, and chromatin stickiness during male meiosis in Anemone tetrasepala (Ranunculaceae). Turk. J. Bot. 37:167–176.
- Podlech D, Dieterle A. 1969. Chromosomenstudien an afghanischen Pflanzen. Candollea. 24:185–243.
- Probatova NS. 2006. Chromosome numbers of vascular plants from nature reserves of the Primorsky Territory and the Amur River basin. Bot. Žurnal (Moscow & Leningrad). 91(7): 1117–1134.
- Raina R, Ashruf M. 1981. In Chromosome number reports LXXII. Taxon. 30(3):707.
- Ranjbar M, Mahmoudi A. 2013a. Chromosome numbers and biogeography of the genus Scutellaria L. (Lamiaceae). Caryologia. 66(3):205–214.10.1080/00087114.2013.821840
- Ranjbar M, Mahmoudi A. 2013b. Cytomorphological study of Scutellaria platystegia (Lamiaceae) in Iran. Feddes Repertorium. 124(1):1–15.
- Ranjbar M, Mahmoudian B. 2015. An overview on cytogenetics of the genus Astragalus subgenus Hypoglottis (Fabaceae). Caryologia. 68(2):109–124.10.1080/00087114.2015.1032571
- Ranjbar M, Karamian R, Nouri S. 2010. Diploid-tetraploid mixoploidy in an interesting new species of Astragalus sect. Incani DC. (Fabaceae) from Iran. Ann. Bot. Fenn. 48(4):343–351.
- Ranjbar M, Karamian R, Nouri S. 2011. Impact of cytomixis on meiosis in Astragalus cyclophyllos Beck (Fabaceae) from Iran. Caryologia. 3(3):256–264.
- Ranjbar M, Pakatchi A, Babataheri Z. 2015. Chromosome number evolution, biogeography and phylogenetic relationships in Salvia (Lamiaceae). Webbia. 70(2):293–312.10.1080/00837792.2015.1057982
- Reese G. 1953. Erganzende Mitteilungen uber die Chromosomenzahlen. Plant Biol. 64:240–255.
- Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, Mayzel J, Chay O, Mayrose I. 2015. The Chromosome Counts Database (CCDB)–a community resource of plant chromosome numbers. New Phytol. 206(1):19–26.10.1111/nph.13191
- Saggoo MI. 1983. Cytomorphological studies on plants of economic importance of Bicarpellatae from India [PhD thesis]. Patiala: Punjabi University. 259 p.
- Saggoo MI, Bir SS. 1982. IOPB chromosome number reports LXXVI. Taxon. 31: 593–595.
- Saggoo MI, Bir SS. 1986. Meiotic studies on some east Himalayan members of family Labiatae. J. Ind. Bot. Soc. 65(3):304–309.
- Salmaki Y, Zarre S, Heubl G. 2012a. The genus Phlomoides Moench (Lamiaceae; Lamioideae; Phlomideae) in Iran: An updated synopsis. Ir. J. Bot. 18(2):207–219.
- Salmaki Y, Zarre S, Ryding O, Lindqvist C, Scheunert A, Bräuchler C, Heubl G. 2012b. Phylogeny of the tribe Phlomideae (Lamioideae: Lamiaceae) with special focus on Eremostachys and Phlomoides: New insights from nuclear and chloroplast sequences. Taxon. 61(1):161–179.
- Sayuri Utsunomiya K, Suely Paglirini M, Borges do Valle C. 2005. Microsporogenesis in tetraploid accessions of Brachiaria nigropedata (Ficalho & Hiern) Stapf (Gramineae). Biocell. 29(3):295–301.
- Scheen AC, Bendiksby M, Ryding O, Mathiesen C, Albert VA, Lindqvist C. 2010. Molecular phylogenetics, character evolution and suprageneric classification of Lamioideae (Lamiaceae). Ann. Mo. Bot. Gard. 97(2):191–217.10.3417/2007174
- Schlarbaum SE, Tsuchiya T. 1984. Cytotaxonomy and phylogeny in certain species of Taxodiaceae. Plant Syst. Evol. 147(1–2):29–54.10.1007/BF00984578
- Shamina NV, Silkova OG, Seriukova EG. 2003. Monopolar spindles in meiosis of intergeneric cereal hybrids. Cell Biol. Int. 27(8):657–664.10.1016/S1065-6995(03)00122-7
- Sheidai M, Koobaz P, Zehzad B. 2003. Meiotic studies of some Avena L. species and populations. J. Sci. I. R. Iran. 14(2):121–131.
- Sheidai M, Alijanpoo B, Khayyami M. 2010. Contribution to cytology of genus Salvia L. (Lamiaceae) in Iran. Caryologia. 63:405–410.
- Sokolovskaya AP, Probatova NS, Rudyka EG. 1986. A contribution to the study of chromosome numbers and geographical distribution of some species of the family Lamiaceae in the Soviet far east. Bot. Žurnal SSSR. 71:195–200.
- Uhrikova A. 1978. In Index of chromosome numbers of Slovakian flora.Part 6. Acta Facultatis Rerum Naturalium Universitatis Comenianae Botanical. 26:1–42.
- Zakirova RO, Nafanailova II. 1988. Chromosome numbers in some species of the Kazakhstan flora. Bot. Žurnal (Moscow & Leningrad). 73:1493–1494.
- Zhukova PG. 1967. Karyology of some plants, cultivated in the Arctic-Alpine Botanical Garden. (In Russian). In: Avrorin NA, editor. Leningrad: Plantarum in Zonam Polarem Transportatio II. p. 139–149.