Abstract
This study is a cytogenetic characterization by karyotyping and determination of the DNA content by flow cytometry of Agave angustifolia Haw. “Cimarron” and “Lineño” that were collected in the southern region of the State of Jalisco, Mexico. These cultivars are particularly important in the production of spirits. A. angustifolia “Cimarron” and “Lineño” were found to be diploids with 2n=2x=60, also, both showed a bimodal karyotype consisting of 10 large + 50 small chromosomes. A. angustifolia “Cimarron” had a karyotype of 42 m+4sm+6st+8t, while the karyotype observed in “Lineño” was of 48 m+2sm+2st+8t. The arm ratio, the proportion of different types of large and small chromosomes, the mean of genome length and the asymmetry index of karyotypes clearly varied between both cultivars, and had a secondary constriction in one pair of the large chromosomes. A. angustifolia “Cimarron” showed a constriction on pair 2 of telocentric chromosomes and “Lineño” on pair 5. The variation between both cultivars is probably due to rearrangements in the large and small chromosomes of the complement. No statistically significant differences were detected in the amount of nuclear 2C DNA = 8.217 pg for “Cimarron” to 8.179 for “Lineño”. The different type of plants displayed 1.7% variation in the 2C DNA content and the mean 2C DNA content was 8.198 pg; 1Cx value = 4.099 pg. The results here reported consist of basic and useful information to set conservation strategies and breeding approaches for Agave angustifolia “Cimarron” and “Lineño”.
Introduction
The genus Agave is endemic to the Americas and includes 200 species. Of these, 75% are found in Mexico, and of which 69% are endemic (García-Mendoza Citation2002, Citation2004, Citation2007). This genus has its domestication and diversity center in Mexico (García-Mendoza Citation2007).
In Mexico, 22 species of Agave are used for mezcal production, most commonly Agave angustifolia Haw, which has the most extensive geographical distribution. Agave angustifolia belongs to the subgenus Agave and is located within the group Rigidae (Gentry Citation1982).
Most of the species of this genus are wild, and constitute an important genetic resource, and are cultivated in different regions of Mexico.
Cytogenetic studies based on meiotic and mitotic chromosome accounts in several taxa of Agave have confirmed its basic chromosome number of x = 30 (Pinkava and Baker Citation1985; Ruvalcaba-Ruiz and Rodríguez-Garay Citation2002; Palomino et al. Citation2005, Citation2008, Citation2010, Citation2015; Moreno Salazar et al. Citation2007).
Diploid species of Agave have bimodal karyotypes, which consist of five pairs of large chromosomes and 25 pairs of small chromosomes (Castorena-Sánchez et al. Citation1991; Vosa Citation2005; Palomino et al. Citation2005, Citation2008, Citation2010, Citation2012a, Citation2015).
In addition, inter and intra-specific variation on chromosome number and karyotypes in Agave species have been reported (Banerjee and Sharma Citation1989; Castorena-Sánchez et al. Citation1991; Vosa Citation2005; Palomino et al. Citation2005, Citation2008, Citation2010, Citation2012a, Citation2015).
Also, DNA content or genome size and molecular markers, such as randomly amplified polymorphism DNA (RAPD), (Rodriguez-Garay et al. Citation2009), amplification fragment length polymorphism DNA (AFLP), (Infante et al. Citation2003; Gil-Vega et al. Citation2006; Abraham-Juárez et al. Citation2009; Sánchez-Teyer et al. Citation2009), ISSR primers, and fluorescent in situ hybridization (FISH) have been used to elucidate the genetic variation in Agave. Gutiérrez-Mora et al. (Citation2010) evaluated the genetic diversity using RAPD markers among plants obtained through sexual and asexual reproduction of Agave angustifolia “Cimarron” and “Lineño”, collected in the southern region of the State of Jalisco, Mexico. In this study, RAPDs allowed a clear separation between the two cultivars, with “Lineño” being more genetically homogeneous than “Cimarron”, putatively due to ancient domestication; furthermore, all analyzed individuals tended to group according to their mode of propagation. Despite this selection process, results obtained through RAPD showed that there is a large genetic variation in these cultivars.
Results of other works also reported the genetic diversity in wild populations of Agave angustifolia in the southern region of Jalisco State, Mexico (Vargas-Ponce et al. Citation2009) and wild populations of A. angustifolia distributed in the State of Sonora, Mexico (Barraza-Morales et al. Citation2006).
On the other hand, the physical mapping of 5S and 18S rDNA (FISH) in several species of Agave was investigated by Gomez-Rodriguez et al. (Citation2013). 5S rDNA loci were located in both arms of a small chromosome pair in each species. The hybridization sites of clonal 18S rDNA were associated with a secondary constriction of a large chromosome pair in each species, being a telocentric chromosome pair number 5 in A. angustifolia “Lineño” and in a telocentric chromosome pair number 2 in “Cimarron”.
Flow cytometry allows a precise and rapid estimation of nuclear DNA content and ploidy levels; it is a method that has a wide range of uses in taxonomy and plant breeding. Moreover, flow cytometry has become increasingly popular, and has been used for genomic analysis in a variety of species (Dolezel et al. Citation2007a).
The present work describes variation in karyotypes and nuclear DNA content by flow cytometry of Agave angustifolia Haw. “Cimarron” and “Lineño”. This information is useful to implement and design programs for maintaining the existing diversity of Agave, and for effective breeding programs to preserve the genetic heritage in these cultivars.
Methods
Plant material
The accession and provenance of two populations of Agave angustifolia “Cimarron” and “Lineño” are listed in Table . Plants were collected in the Southern region of the State of Jalisco, México (Municipality of Tolimán). Plants were transported to the Botanical Garden (Instituto de Biología, UNAM, planted in pots containing a mixture of organic soil, sand and vermiculite and kept under greenhouse conditions). Voucher specimens were deposited at the GUADA herbarium (Universidad Autónoma de Guadalajara, Guadalajara, Jalisco, México).
Table 1. Provenance and karyotype analysis in diploid Agave angustifolia cultivars (2n = 2x = 60).
Mitotic chromosome counts
Mitotic cells (18 to 16) at metaphase stage were observed from eight to six individual plants of each cultivar in order to evaluate chromosome numbers (2n) and karyotypes.
Elongating secondary root tips were treated with 0.002 M 8-hydroxy-quinoline for 6 h at 18°C in darkness. Later, the root tips were fixed in fresh Farmer solution (three parts absolute ethanol: one part glacial acetic) for 24 h. The root tips were hydrolyzed with 1 N hydrochloric acid for 15 min at 60°C, and transferred to Schiff reagent for 1 h, and then to 1.8% propionic orcein to stain chromosomes. Slides were prepared and frozen with dry ice (Conger and Fairchild Citation1953) and mounted in Canada balsam. Twelve of the best cells of each cultivar were photographed using an Axio Vision Rel. 4.7 camera in a Zeiss photomicroscope III (Göttingen, Germany).
Karyotype analysis
A negative film was used to draw and measure chromosomes arms and the total genome length. The centromere position was obtained following Levan et al. (Citation1964); arm ratio (r = long arm/short arm) was calculated for each chromosome (Table ). Chromosome homology was assigned according to similarities in length and centromere position. In addition, secondary constrictions were useful to distinguish homologous pairs in both populations. Ideograms were constructed according to the arm ratio of the chromosomes, and then grouped in metacentric (m), submetacentric (sm), subtelocentric (st) and telocentric (t) chromosomes. The number of homologous chromosomes was sequentially assigned following chromosome length, for a total number of 30. The index of asymmetry (TF%) was obtained following the procedure of Gupta and Gupta (Citation1978).
Table 2. Arm ratio (r) of homologous chromosome pairs of Agave angustifolia cultivars.
Estimation of nuclear DNA content
Six to eight plants were used for the nuclear DNA content estimation, using flow cytometry, in the two studied populations of Agave angustifolia. Three replicates for each individual plant were analyzed. Maize (Zea mays cv. CE-777) with 2C DNA = 5.4333 pg (Dolezel et al. Citation2007b) was used as an internal standard for diploid cultivar plants. Samples were prepared according to Dolezel et al. (Citation2007b). Briefly, 100–120 mg of A. angustifolia internal basal leaf tissue and 30 mg of maize (Zea mays cultivar CE-777) with 2C DNA = 5.433 pg were simultaneously chopped with a razor blade in a Petri dish containing 1.5 ml of 0.1 M citric acid and 0.5% Tween 20. Chopped material was filtered through a 50 μm nylon mesh and incubated for 15 min at room temperature. The nuclei in the filtrate were pelleted by centrifugation (90 g for 3 min), suspended in 1 ml of the citric acid/Tween 20 solution, and incubated for 15 min at room temperature. Then, 2 ml of 0.4 M Na2HPO4 was added and the suspension was supplemented with 125 μl of propidium iodide and RNase to a final concentration of 50 μg ml−1. The mixture was then analyzed by flow cytometry.
Flow cytometric analysis
The flow cytometric estimation of the nuclear DNA content was performed using a Partec CY Flow SL cytometer (Partec, Munster, Germany). Calibration green beads (Partec) were used to align the flow cytometer and for checking its linearity.
The gain recorded attributed to the instrument was adjusted so that the peak representing G1 nuclei of Zea mays was positioned on channel 50 of the 250-channel linear scale. At least 10,000 nuclei were analyzed for each sample. Peak means, areas and coefficient of variation were calculated using the DPAC software (Partec). Coefficients of variation were less than 5% assuring accurate measurements (Dolezel et al. Citation2007b). Nuclear genome size was then calculated according to Dolezel et al. (Citation2007b) by using the formula:(1)
where A = agave 2C nuclear DNA content (pg); B = agave G0/G1 peak mean; C = internal standard G0/G1 peak mean; D = 2C DNA content internal standard (pg); 1 picogram (pg) = 978 megabase pairs (Mbp) (Dolezel et al. Citation2007b).
Statistical analyses
The simultaneous differences among the plants of Agave angustifolia “Cimarron” and “Lineño”, in terms of the mean length of homologous chromosome pairs, were evaluated according to 30 nested and unbalanced ANOVA analyses performed for each homologous chromosome pair. An unbalanced bifactorial ANOVA design with 34 cells was applied to 6–8 plants of each cultivar to evaluate differences in the total diploid genome length and asymmetry index (TF %), where the main factor was the cultivar of Agave angustifolia, and the individual differences within each type of plants was the random effect.
Differences in 2C DNA content in pg and Mbp for each cultivar were evaluated according to a complete nested unbalanced analysis of variance (ANOVA). The first level of analysis corresponded to the different cultivar of plants; the second level was the analysis of the differences among individuals in each cultivar; and the third level was the comparison of replicates within each individual plant. Additionally, restricted maximum likelihood estimation (REML) was applied to data analysis; in both cases, variance homogeneity was tested. The Shapiro–Wilk procedure was used to test normality for the studentized residual from all the ANOVA analyses. In those cases where studentized residuals were not normally distributed, a Box–Cox transformation was performed. If results of these analyses were significant, a Tukey–Kramer test for means differences was applied. All statistical analyses were performed using the JMP version 11.0 software (SAS Institute, Cary, NC, USA).
Results
Chromosome number
Chromosome number in well-spread mitotic metaphase cells of Agave angustifolia “Cimarron” and “Lineño” from Tolimán Jalisco State, Mexico, were diploids with 2n = 2x = 60 (Tables and , Figures (a), (b), (a), (b)). All plants of the studied populations were euploid with the basic chromosome number of x = 30.
Figure 1. Chromosome of diploid Agave angustifolia “Cimarron” (2n = 2x = 60) from Toliman, Jalisco, Mexico: (a) mitotic metaphase; (b) idiogram. Numbers indicate homologous large chromosome pair with secondary constriction. t: telocentric chromosome, st: subtelocentric chromosome, sm: submetacentric chromosome and m: metacentric chromosome. Bar: 10 μm.
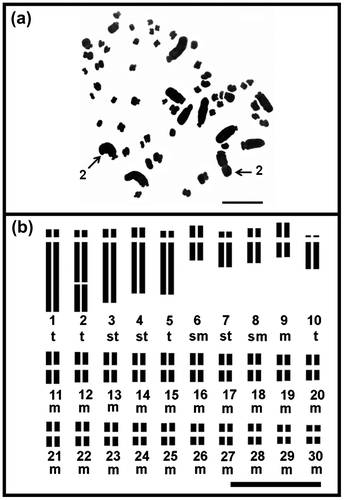
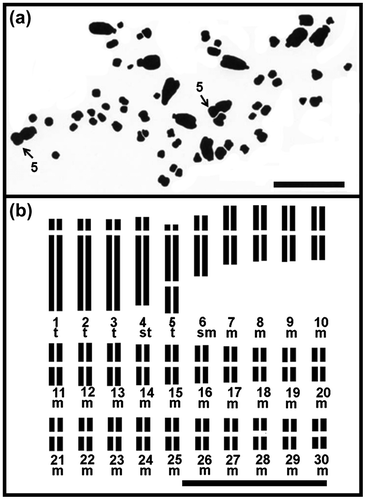
Figure 2. Chromosome of diploid Agave angustifolia “Lineño” (2n = 2x = 60) from Toliman, Jalisco, Mexico: (a) mitotic metaphase; (b) idiogram. Numbers indicate homologous large chromosome pairs with secondary constriction. t: telocentric chromosomes, st: subtelocentric chromosome, sm: submetacentric chromosome and m: metacentric chromosome. Bar: 10 μm.
Karyotype analysis
Two different structural diploid karyotypes were observed in Agave angustifolia “Cimarron” and “Lineño” (Tables ), and no previous karyotype determination has been reported for these cultivars.
Table 3. Chromosome analysis of Agave angustifolia cultivars.
Table 4. Results of Tukey–Kramer test on mean length of homologous monoploid chromosome set (x = 30) of Agave angustifolia cultivars.
The mean of the range of the chromosome length for the diploid Agave angustifolia “Cimarron” and “Lineño” is shown in Table . Agave angustifolia “Cimarron” showed a karyotype of 42 m+4sm+6st+8t. A. angustifolia “Lineño” displayed a karyotype of 48 m+2sm+2st+8t, and both cultivars showed one pair of large telocentric chromosome with a secondary constriction.
The five pairs of large chromosomes had the highest values for arm ratio (r) in both cultivars (Table ). The existence of three different chromosome types of large chromosomes in these two cultivars which differ in size, centromere position, and secondary constriction, were evident and corresponded to: large telocentric chromosome with a secondary constriction (Lt*); large telocentric chromosome (Lt), and large subtelocentric chromosome (Lst). The secondary constriction was located on pair 2 of A. angustifolia “Cimarron”, and on pair 5 in “Lineño”. The five pairs of large chromosomes had the highest value of arm ratio (r) in the karyotype of both cultivars. The (r) parameter was the least variable in the group of 25 small chromosomes (Tables , Figures ).
A. angustifolia “Cimarron” had telocentric chromosomes in pairs number 1, 2 and 5 with an arm ratio r = 16.19, 10.37 (Lt*) and 10.25 respectively. In contrast “Lineño” had telocentric chromosomes in pairs numbers 3, 5, 2 and 1 with smaller arm ratio values of r = 9.00, 7.89 (Lt*), 7.25 and 7.12, respectively.
Regarding the group of small chromosomes, four types were found: small telocentric chromosome (St); small subtelocentric chromosomes (Sst); small submetacentric chromosomes (Ssm) and small metacentric chromosome (Sm). A variation in the proportion of small chromosome type was also observed in the complements of “Cimarron” and “Lineño” (Tables , Figures (a), (b), (a), (b)).
The position of a secondary constriction was displayed in different pairs of large chromosomes in the complement of the two cultivars. “Cimarron” showed it on pair 2, and “Lineño” on pair 5 of telocentric chromosomes of the large group of chromosomes of their complement.
The mean length of the large chromosomes was different in both cultivars. Highly significant differences were found in pairs 2 (p = 0.0107), 3 (p = 0.0196), 4 (p = 0.0046) and 5 (p = 0.0022) of long chromosomes while in the group of small chromosomes differences were found on pairs 12 (p = 0 089), 13 (p = 0.0017), 14 (p = 0.0018), 23 (p = 0.0473), 24 (p = 0.0338), 26 (p = 0.48), 29 (p = 0.0201), and 30 (p < 0.0001) (Table ).
Regarding the 25 small chromosomes, the value of arm ratios (r) was less variable than those of large chromosomes; “Cimarron” had st chromosome pair number 7 with an arm ratio r = 3.42. In contrast “Lineño” showed sm chromosome pair number 6 with r = 2.66, (Tables ).
All the analyzed plants had a bimodal karyotype with five pairs of large homologous chromosomes and 25 pairs of small homologous chromosomes (Tables , Figures ).
Figure 3. Mean length of homologous chromosome pairs (x = 30) in Agave angustifolia “Cimarron” (+) and “Lineño” (x). Diploid populations (2n = 2x = 60) from Toliman, Jalisco State, Mexico.
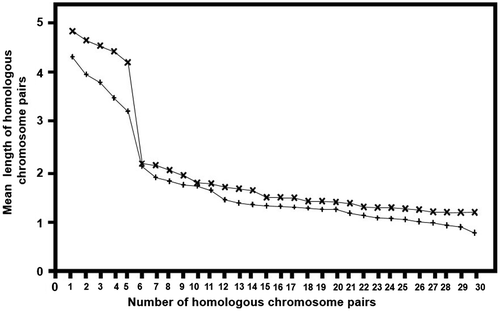
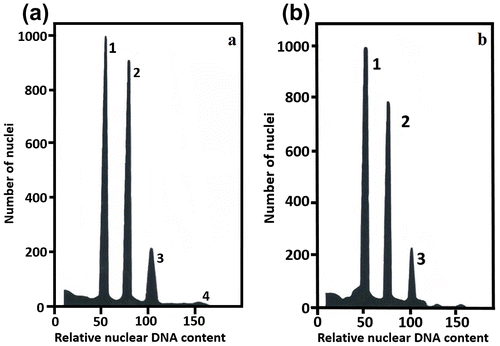
Figure 4. Estimation of nuclear DNA content in Agave angustifolia “Cimarron” and “Lineño” using flow cytometry. (a) Simultaneous analysis of nuclei isolated from A. angustifolia “Cimarron” (2n = 2x = 60) and Zea mays cv. CE-777, used as standard for diploid A. angustifolia “Cimarron”. Peaks 1 and 3 represent G1 and G2 nuclei of Z. mays. Peaks 2 and 4 represent G1 and G2 of A. angustifolia “Cimarron”. (b) Simultaneous analysis of nuclei represents G1 and G2 nuclei of Z. mays. Peaks 2 and 4 represent G1 and G2 of diploid A. angustifolia “Lineño” (2n = 2x = 60) and Z. mays cv. CE-777, used as standard for “Lineño”.
The total diploid genome length was 102.69 μm for “Cimarron”, and 121.12 μm for “Lineño”. No significant differences among them were found (p>0.325; Table ).
The value of asymmetry index (TF %) was not significant different in the populations of Agave angustifolia “Cimarron” and “Lineño” studied (p>0.223; Table ).
Genome size variation in Agave angustifolia “Cimarron” and “Lineño”
The nuclear DNA content analysis revealed that the 2C DNA content estimated for two Agave angustifolia diploid cultivars had no significant differences (p<0.4543) and ranged from 8.217 pg (1Cx = 4013 Mbp) in plants of “Cimarron” to 8.179 pg (1Cx = 4002 Mbp) in “Lineño”, showing a difference of 1.7%. The mean 2C DNA content was 8.198 pg; 1Cx value = 4.009 pg. No nuclei had DNA content higher than 4C (Figure (a), (b)). These results indicate the absence of an endopolyploidy pattern in both of the Agave angustifolia cultivars (Table , Figure (a), (b)).
Table 5. Nuclear DNA content and genome size from Agave angustifolia cultivars.
Discussion
Diploid species of genus Agave have been reported with chromosome numbers of 2n = 2x = 60; x = 30 (Pinkava and Baker Citation1985; Ruvalcaba-Ruiz and Rodríguez-Garay Citation2002). These results are concordant with the data reported here. The findings pertaining to variation in r values for large and small chromosomes of karyotypes of all plants of A. angustifolia “Cimarron” and “Lineño” studied, clearly document changes and rearrangements that are part of the chromosome evolution process of Agave species. These results are evidenced by significant changes in the chromosome structure and in karyotype variation of all the studied plants.
The bimodal karyotype found in Agave angustifolia “Cimarron” and “Lineño” has been reported in other species of Agave (Gómez-Pompa et al. Citation1971; Banerjee and Sharma Citation1988, Citation1989; Finch and Osborne Citation1990; Vosa Citation2005; Moreno-Salazar et al. Citation2007; Palomino et al. Citation2005, Citation2008, Citation2010, Citation2012a, Citation2015); and its bimodal karyotype results from an evolutionary process known as orthoselection (Brandham and Doherthy Citation1998).
All individual plants analyzed in each cultivar of Agave angustifolia showed the same karyotype. A. angustifolia “Cimarron” had a karyotype of 42 m+4sm+6st+8t, while A. angustifolia “Lineño” displayed a karyotype of 48 m+2sm+2st+8t.
When analyzing the parameter r (r = long arm/short arm) in the chromosomes of A. angustifolia “Cimarron” and “Lineño”, significant changes were evident in some chromosome pairs. In “Cimarron”, the group of large chromosomes showed remarkable changes in chromosome pairs 2, 3 and 5, which were telocentric, subtelocentric and telocentric chromosomes respectively; showing the double constriction in pair 2 of telocentrics.
In “Lineño” four telocentric pairs and one subtelocentric pair were found showing the double constriction on pair 5. When comparing the complement of the studied populations, the existence of three different chromosome types of large chromosomes, which differ in size, centromere position, and secondary constriction (sc), were evident in the karyotype of “Cimarron” and “Lineño” and corresponded to: large telocentric chromosomes with a secondary constriction (Lt*); large telocentric chromosomes (Lt); and large subtelocentric chromosome (Lst). This type of chromosome varied in number in the two cultivars studied. In “Cimarron”, two pairs of long chromosomes t and two pairs st were observed. On the other hand, “Lineño” showed three t pairs and just one pair of st (Tables , Figures ).
In the group of small chromosomes of “Cimarron”, differences in the r value were evident in pairs 7, 8 and 10 and chromosomes were subtelocentrics (st), submetacentrics (sm) and telocentrics (t), respectively. In “Lineño”, differences in the r value were evident in pair 6 that corresponded to submetacentrics (sm) (Tables , Figures ).
The higher values of arm ratio were found in the group of the five large chromosomes and the smaller value of arm ratio in the 25 small chromosomes; this gave evidence of the changes and rearrangements in the large and small chromosomes of both cultivars. Different structural rearrangements have been reported in Agave stricta as inversion heterozygosity and subchromatid exchange (Brandham Citation1969).
Gomez-Rodríguez et al. (Citation2013) investigated the physical mapping of 5S and 18S rDNA by fluorescent in situ hybridization (FISH) in several species of Agave, and showed that the number of sites of rDNA were constant among all species under study. The hybridization sites of clones 18S rDNA were associated with a secondary constriction of a large chromosome pair in each species. In Agave angustifolia “Cimarron” and “Lineño”, a secondary constriction was observed in pairs 2 and 5 of telocentric chromosomes respectively of the large chromosome group of the complement of these cultivars. The results obtained by these authors were similar to the results obtained in this work. Furthermore, the 5S rDNA loci were located in both arms of small chromosome pair number 11 in both cultivars (Gómez-Rodriguez et al. Citation2013).
Genome size variation and ploidy level in Agave angustifolia “Cimarron” and “Lineño”
Palomino et al. (Citation2003) reported the genome size of eight cultivars (2n=2x=60) of Agave tequilana Weber. The average 2C DNA content of diploid cultivars ranged from 8. 304 pg for “Lineño” to 8.517 pg for “Pata de Mula”; thus, only small differences (2.5%) in DNA content were detected. A Tukey test showed that the differences in the amount of 2C DNA in diploid cultivars were not statistically significant; however, high significant differences (p<0.001) among the DNA content of diploid cultivars were found when compared with polyploid cultivars of A. tequilana. The ICx (DNA amount of monoploid chromosome set) calculated for diploid cultivars was 4.202 pg DNA.
The genome size has been studied in 36 species, 12 cultivars and one subspecies of Agave (Palomino et al. Citation2007). The authors observed that 2C DNA content for diploid taxa were around 8.0 pg. In diploid taxa 2C DNA content ranges from 6.0 pg in Agave fourcroydes (Banerjee and Sharma Citation1988) to 9.6 pg in A. desertii (Bennet and Smith Citation1991) with around 60% variation in genome size.
The DNA content of A. angustifolia diploid “Cimarron” and “Lineño” is indicative of a medium-sized genome according to the range defined for angiosperms from the smallest in Genlisea margaretae (1C = 0.0648 pg) to the largest in Paris japonica (1C = 152.23 pg) genomes thus far reported, extending the range found in angiosperm to nearly 2400-fold (Bennett and Leitch Citation2011).
Conclusions
In summary, inter- and intraspecific karyotypic variation has been reported in Agave angustifolia “Cimarron” and “Lineño” and in several other species, varieties, and agronomically important cultivars of species of Agave (Castorena-Sánchez et al. Citation1991; Palomino et al. Citation2005, Citation2008, Citation2012a, Citation2015; Moreno-Salazar et al. Citation2007). Variation in karyotypes and genome size has been found in other species with both systems of reproduction sexual and vegetative, e.g. Lycoris chinensis (Liu et al. Citation2012); Echeandia spp. (Palomino et al. Citation2012b); and Nothoscordum bivalve (Palomino et al. Citation1992). Species with sexual reproduction, e.g. Cichorium intybus (Bernardes et al. Citation2013), Lysimachia mauritiana (Kono et al. Citation2012), Tulipa species (Abedi et al. Citation2015), and Veronica species (Albach and Greilhuber Citation2004), also showed intraspecific variation in their karyotypes and genome size. This variation can be attributed to the plasticity of each species to adapt to different environmental situations and to increase their fecundity during asexual reproduction (Marcucci and Tornadore Citation1997).
This type of research on the genome size of Agave angustifolia “Cimarron” and “Lineño” is necessary to better design and apply programs to enhance within-species diversity. Finally, the results of this study will be useful for future programs focused on the analysis of plant genomes, including construction of DNA libraries and physical mapping in these important Agave species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was partially supported by Red de Agavaceas-Servicio Nacional de Inspección y Certificación de Semillas-México [project number 50349], Jardín Botánico, Instituto de Biología, Universidad Nacional Autónoma de México (IBUNAM) and Red temática mexicana aprovechamiento integral sustentable y biotecnología de los agaves (AGARED).
Acknowledgments
We thank Dr A. García-Mendoza for his help in the identification of plants used in this research, M. Ladd for her technical assistance and Felipe Villegas for the art work.
References
- Abedi R, Babel A, Karimzadeh G. 2015. Karyological and flow cytometric studies of Tulipa (Liliaceae) species from Iran. Plant Syst Evol. 301:1473–1484.10.1007/s00606-014-1164-z
- Abraham-Juárez MJ, Ramírez-Malagón R, Gil-Vega KC, Simpson J. 2009. Análisis AFLP de la variabilidad Genética en tres formas de reproducción de Agave tequilana. Rev Fitotec Mex. 32(3):171–175.
- Albach DC, Greilhuber J. 2004. Genome size variation and evolution in Veronica. Ann Bot. 94:897–911.10.1093/aob/mch219
- Banerjee S, Sharma AK. 1988. Structural differences of chromosomes in diploid Agave. Cytologia. 53(2):415–420.10.1508/cytologia.53.415
- Banerjee S, Sharma AK. 1989. Structure and behaviour of chromosomes in four different species of Agave. Cytologia. 54(4):667–672.10.1508/cytologia.54.667
- Barraza-Morales A, Sanchez FL, Robert M, Esqueda M, Gardea A. 2006. Variabilidad genética en Agave angustifolia Haw. de la Sierra sonorense determinada con marcadores AFLP. Rev Fitotec Mex. 29(1):1-8.
- Bennett MD, Smith JB. 1991. Nuclear DNA amount in angiosperms. Philos T Roy Soc S B. 334(1271):309–345.10.1098/rstb.1991.0120
- Bennett MD, Leitch IJ. 2011. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot. 107:467–590.10.1093/aob/mcq258
- Bernardes ECS, Benko-Iseppon AM, Vasconcelos S, Carvalho R, Brasileiro-Vidal AC. 2013. Intra- and interespecific chromosome polymorphisms in cultivated Chichorium L. species (Asteraceae). Genet Mol Biol. 36(3):357–364.10.1590/S1415-47572013005000025
- Brandham PE. 1969. Inversion heterozygosity and sub-chromatid exchange in Agave stricta. Chromosoma (Berlin). 26(3):270–286.10.1007/BF00326522
- Brandham PE, Doherty MJ. 1998. Genome size variation in the Aloaceae, an angiosperm family displaying karyotypic orthoselection. Ann Bot. 82(1) ( Supplement A):67-73. 10.1006/anbo.1998.0742
- Castorena-Sánchez I, Escobedo RM, Quiroz A. 1991. New cytotaxonomical determinants recognized in six taxa of Agave in the sections Rigidae and Sisalanae. Can J Bot. 69(6):1257–1264.10.1139/b91-163
- Conger DD, Fairchild LM. 1953. A quick-freeze method for making smear slides permanent. Stain Technol. 28(6):281–283.10.3109/10520295309105555
- Dolezel J, Greilhuber J, Suda J. 2007a. Flow cytometry with plants: an overview. In: Dolezel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Weinheim (Germany): Wiley-VCH. p. 41–65.10.1002/9783527610921
- Dolezel J, Greilhuber J, Suda J. 2007b. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2(9):2233–2244.10.1038/nprot.2007.310
- Finch RA, Osborne JF. 1990. Chromosome numbers and DNA amounts in Agave variants. East Afric Agric Forest J. 55(4):213–218.
- García-Mendoza A. 2002. Distribution of Agave (Agavaceae) in México. Cact Succ J (USA). 74(4):177–187.
- García-Mendoza A. 2004. Agaváceas. In: García-Mendoza AJ, Ordoñez MJ, Briones-Salas M, editors. Biodiversidad de Oaxaca. Instituto de Biología, UNAM-Fondo Oaxaqueño para la Conservación de la Naturaleza-World Wildlife Fund México. p. 159–169.
- García-Mendoza A. 2007. Los agaves de México. Ciencias. Universidad Nacional Autónoma de México, Distrito Federal, México. 87:14–23.
- Gentry HS. 1982. Agaves of Continental North America. Tucson (AZ): The University of Arizona Press.
- Gil-Vega K, Díaz C, Nava-Cedillo A, Simpson J. 2006. AFLP analysis of Agave tequilana varieties. Plant Sci. 170(4):904–909.10.1016/j.plantsci.2005.12.014
- Gómez-Pompa A, Villalobos-Pietrini R, Chimal A. 1971. Studies in the Agavaceae. 1. Chromosome morphology and number of seven species. Madroño. 21:208–221.
- Gomez-Rodriguez VM, Rodriguez-Garay B, Palomino G, Martínez J, Barba-González R. 2013. Physical mapping of 5s and 18s ribosomal DNA in three species of Agave (Asparagales, Asparagaceae). Comp Cytogenet. 7(3):191–203.
- Greilhuber J, Dolezel J, Lysak M, Bennett MD. 2005. The origin, evolution and proposed stabilization of the terms genome size and C-value to describe nuclear DNA content. Ann Bot. 95(1):255–260.10.1093/aob/mci019
- Gupta R, Gupta PK. 1978. Karyotypic studies in the genus Crotalaria Linn. Cytologia. 43(2):357–369.10.1508/cytologia.43.357
- Gutiérrez-Mora A, Arana-Gutiérrez JPR, Tapia-Campos E, Rodríguez-Garay B. 2010. Molecular analysis of sexual and asexual genetic variation of two sympatric Agave angustifolia varieties. J PACD. 12:155–165.
- Infante D, González G, Peraza-Echeverría L, Kleb-Llanes M. 2003. Asexual genetic variability in Agave fourcroydes. Plant Sci. 164:223–230.10.1016/S0168-9452(02)00404-1
- Kono Y, Chung KF, Chen CH, Hoshi Y, Setoguchi H, Chou CH. 2012. Intraspecific karyotypicpolymorphism is highly concoedant with allozyme variation in Lysimachia mauritiana (Primulaceae; Myrsinoideae) in Taiwan: implications for the colonization history and dispersal patterns of coastal plants. Ann Bot. 110(6):1119–1135.10.1093/aob/mcs192
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosome. Hereditas. 52(2):201–220.
- Liu K, Zhou SB, Huang YJ, Tang CF, Zhang D, Huang ZZ. 2012. Chromosomal variation and evolution in Lycoris (Amarillidaceae) I. Intraspecific variation in the karyotype of Lycoris chinensis Traub Plant Syst Evol 298(8):1493-1502.10.1007/s00606-012-0652-2
- Marcucci R, Tornatore N. 1997. Intraspecific variation of Allium roseum L. (Alliaceae). Webbia. 52(1):137–154.10.1080/00837792.1997.10670636
- Moreno-Salazar SF, Esqueda M, Martínez J, Palomino G. 2007. Tamaño del genoma y cariotipo en Agave angustifolia y A. rhodacantha de Sonora, México. Rev Fitotec Mex 30(1): 13-23.
- Palomino G, Romo V, y Ruenes R. 1992. Fisiones céntricas en cromosomas metacéntricos de Nothoscordum bivalve (L.) Britt. (Alliaceae) de México. Bol Soc Bot Mex. 52:121–124.
- Palomino G, Dolezel J, Méndez I, Rubluo A. 2003. Nuclear genome size analysis of Agave tequilana Weber. Caryologia. 56(1):37–46.10.1080/00087114.2003.10589305
- Palomino G, Martínez J, Méndez I. 2005. Citotipos en Agave angustifolia Haw. determinados por citometría de flujo y análisis de sus cariotipos. Rev Int Cont Amb 21(Suppl. 1):49-54.
- Palomino G, Martínez J, Méndez I. 2007. Variación inter e intraespecífica en especies de Agave por citometría de flujo y análisis de sus cromosomas. In: Colunga-García MP, Larqué S, Eguiarte LE, Zizumbo-Villareal D, editors. En lo ancestral hay futuro: del tequila, los mezcales y otros agaves. Mérida, Yucatán. México: CICY, CONACYT, CONABIO y SEMARNAT. p. 41-64.
- Palomino G, Martínez J, Méndez I. 2008. Karyotype studies in cultivars of Agave tequilana Weber. Caryologia. 61(2):144–153.
- Palomino G, Martínez J, Méndez I. 2010. Análisis del tamaño del genoma y cariotipo de Agave aktites Gentry (Agavaceae). Rev Mex Biodiv. 81(3):655–662.
- Palomino G, Martínez J, Cepeda-Cornejo V, Pimienta-Barrios E. 2012a. Nuclear genome size and cytotype analysis in Agave cupreata Trel & Berger (Agavaceae). Caryologia. 65(4):281–294.10.1080/00087114.2012.752915
- Palomino G, Martínez J, Barba –González R, Méndez I, Rodríguez-Garay B. 2012b. Mexican geophytes III. Cytotypes and meiotic behavior in Mexican populations of species of Echeandia (Anthericaceae). In: Floric Orn Biotech. 6 (Special Issue 1):140-152. Global Science Books.
- Palomino G, Martínez J, Méndez I, Cepeda-Cornejo V, Barba –González R, Rodríguez-Garay B. 2015. Nuclear genome size and cytotype analysis in Agave parviflora Torr subsp. flexiflora Gentry & Berger (Asparagales, Asparagaceae). Caryologia 68(3):159-168. 10.1080/00087114.2015.1032575
- Pinkava DJ, Baker MA. 1985. Chromosome and hybridization studies of Agaves. Desert Plants. 7:93–100.
- Rodríguez-Garay B, Lomelí-Sención JA, Tapia-Campos E, Gutierrez-Mora A, García-Galindo J, Rodríguez-Domínguez JM, Urbina-López D, Vicente-Ramírez I. 2009. Morphological and molecular diversity of Agave tequilana Weber var. azul and Agave angustifolia Haw. Var Lineño Ind Crop Prod. 29:220–228.10.1016/j.indcrop.2008.05.007
- Ruvalcaba-Ruiz D, Rodríguez-Garay B. 2002. Aberrant meiotic behavior in Agave tequilana Weber var. azul. BMC Plant Biol. 2:10.10.1186/1471-2229-2-10
- Sánchez-Teyer LF, Moreno-Salazar S, Esqueda M, Barraza A, Robert ML. 2009. Genetic variability of wild Agave angustifolia populations based on AFLP: A basic study for conservation. J Arid Environ. 73:611–616.10.1016/j.jaridenv.2009.01.008
- Vargas-Ponce O, Zizumbo-Villareal D, Martínez-Castillo J, Coello-Coello J, Colunga-GarcíaMarín P. 2009. Diversity and structure of landraces of Agave grown for spirits under traditional agricultura: A comparison with wild populations of A. angustifolia (Agavaceae) and commercial plantations of A. tequilana. Am J Bot. 96(2):448–457.10.3732/ajb.0800176
- Vosa CG. 2005. On chromosome uniformity, bimodality and evolution in the tribe Aloineae (Asphodelaceae). Caryologia. 58:83–85.10.1080/00087114.2005.10589437
