Abstract
In this study, karyotypes of four species of the genus Quercus (Quercus infectoria subsp. boissieri (Reuter) O. Schwarz, Q. pubescens Willd., Q. robur L. and Q. vulcanica (Boiss & Heldr. Ex) Kotschy) sampled in Turkey were determined. Each analysed species had the same chromosome number with 2n = 24, all of them were metacentric. Although the chromosome numbers of studied taxa were compatible with previous studies, some differences were determined at the intra-specific level. The endemic species Q. vulcanica were analysed for the first time in this study.
Introduction
The Fagaceae family, which represents woody plants distributed throughout the Northern Hemisphere, contains approximately 1000 species. There are nine recognized genera in the family: Castanea L., Castanopsis Spach., Chrysolepsis Hjelmquist, Colombobalanus Nixon & Crepet, Fagus L., Formanodendron Nixon & Crepet, Lithocarpus Blume, Quercus L. and Trigonobalanus Forman (Borgardt and Kathleen Citation1999; Doğan et al. Citation2000).
The genus Quercus, with high diversity and number of species, dominates many regions of temperate, tropical and subtropical forests of Europe and North America (Nixon Citation2002). The genus contains more than 500 species distributed in the temperate zone in the Northern Hemisphere (Govaerts and Frodin Citation1998; Olfat and Pourtahmasi Citation2010; Maryam Ardi et al. Citation2012).
Turkey is among the richest countries in the number of Quercus taxa, with 18 species belonging to three sections, Quercus, Cerris and Ilex (Yaltirik Citation1984).
Section Quercus has the greatest number of species and widest distribution in the world. In Turkey, most species belonging to the genus Quercus are in section Quercus: Q. pontica C. Koch, Q. robur L., Q. hartwissiana Steven, Q. macranthera subsp. sysprensis (C. Koch) Menitsky, Q. frainetto Ten., Q. petraea (Mattuschka) Liebl., Q. vulcanica (Boiss & Heldr. Ex) Kotschy, Q. infectoria Oliver, Q. pubescens Willd. and Q. virgiliana Ten. (Yaltirik Citation1984).
Oak taxonomy is not completely clear. Oaks exhibit the highest genetic variation among all woody plant species (Kremer and Petit Citation1993). Oaks are geographically widespread species and many species grow in mixed populations where hybridization is common (Bacilieri et al. Citation1996). Reproductive barriers between oak species are weak (Manos et al. Citation1999; Borazan and Babaç Citation2003) and extensive hybridization may occur between species in the same section or group within the genus.
Taxonomic problems are also increased by introgression, ecological adaptation and the use of inconsistent morphological characters as species descriptors (Denk and Grimm Citation2010; Simeone et al. Citation2013).
In spite of the many recent molecular studies on oak species, cytological investigations are still insufficient (Ohri and Ahuja Citation1990; Zoldo et al. Citation1998; Ribeiro et al. Citation2011); this is particularly the case for tree species with small chromosomes, as oak species do (Chokchaichamnankit et al. Citation2008).
This study aims to identify three oak species (Quercus infectoria subsp. boissieri, Q. pubescens and Q. robur) with similar distributions in Turkey and Europe, and to determine the relationships among the oak species studied and Europe oaks. At the same time, an oak species endemic to Turkey (Q. vulcanica) was examined for the first time in detail.
Chromosome numbers, karyotypes and morphometric parameters, including the haploid complement, intrachromosomal asymmetry (A1), and interchromosomal asymmetry (A2), were identified for different populations in Turkey of Quercus infectoria subsp. boissieri, Q. pubescens, Q. robur and the endemic species Q. vulcanica.
Intrachromosomal asymmetry index (A1) expresses the arm ratio of each pair of homologous chromosomes, and the interchromosomal asymmetry index (A2) gives an idea of the asymmetry caused by the different length of the chromosomes. In this study by using the Romero Zarco asymmetry indices (A1 and A2), it is aimed to determine the more asymmetric karyotypes among the populations.
Materials and methods
Plant samples were collected from different localities in Turkey. The locations and species are presented in Table . Acorns of each studied species were germinated at 4°C for somatic chromosomal preparations. Root tips obtained from acorns were taken and pretreated in α-monobromonaphthalene for 14–16 h at 4°C. After first treatment, fixation was done with Carnoy solution overnight. Fixed root tips were transferred to 70% alcohol and stored in a refrigerator until analyses. Afterwards, hydrolysis was done with 1 N HCl at 60°C for 13 min. Finally the root tips were stained with Feulgen for 2 h and then squashes were made with 2% aceto-orcein. Preparations containing metaphase chromosomes were frozen in liquid nitrogen to make permanent with Entellan. In each mitotic metaphase, the arm length of each chromosome was measured at least five plates.
Table 1. Species names, localities and chromosome numbers of studied species.
Chromosomes for each species were classified according to the nomenclature of Levan et al. (Citation1964) and Stebbins (Citation1971).
The relative lengths, karyotypic description and length range of all studied species were calculated in addition to the karyotype asymmetry parameters: intrachromosomic asymmetric index (A1) and interchromosomic asymmetric index (A2), following Romero Zarco (Citation1986).
Results and discussion
Materials for cytotaxonomic study were obtained from different localities in Turkey (Table ). We determined detailed chromosome measurements and numbers of four taxa from the genus Quercus for the first time in Turkey (Tables and ). Analysis of somatic metaphase plates for Quercus taxa, namely Quercus infectoria subsp. boissieri, Q. pubescens, Q. robur and Q. vulcanica, identified 2n = 24 chromosomes in all studied species (Tables and , Figures and ).
Table 2. Karyotypic descriptions, length ranges and other morphometric parameters of studied Quercus taxa.
Figure 1. Somatic chromosomes of (a) Q. infectoria subsp. boissieri; (b) Q. pubescens; (c) Q. robur; (d) Q. vulcanica.
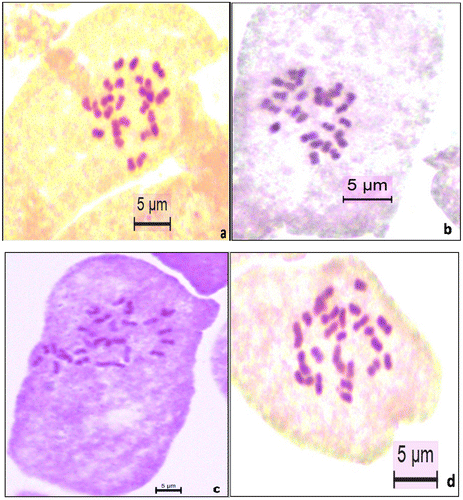
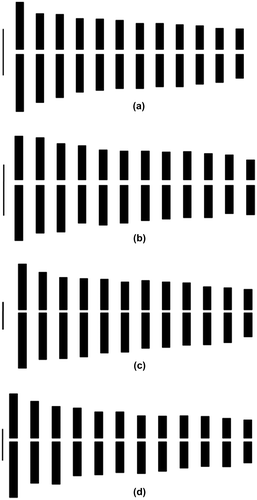
Figure 2. Idiograms of (a) Q. infectoria subsp. boissieri; (b) Q. pubescens; (c) Q. robur; (d) Q. vulcanica (bar = 1 μm).
The following features of the four Quercus taxa examined were observed in this study:
| (1) | The chromosome number of Quercus infectoria subsp. boissieri was found to be 2n = 24. All chromosomes analysed were metacentric. Among of the studied species, Quercus infectoria subsp. boissieri has the smallest chromosome set after Q. pubescens. Chromosomal length range was 1.02–2.35 μm and haploid complement value was 17.89 μm. | ||||
Quercus infectoria consists of two subspecies (Q. infectoria subsp. infectoria and Q. infectoria subsp. boissieri) in Turkey and has the widest distribution area, especially in the west, south and south-east regions of Turkey. Q. infectoria subsp. infectoria has a more limited distribution area compared with Q. infectoria subsp. boissieri in Turkey.
Chromosome number, chromosome types and morphometric parameters including other detailed chromosome measurements of Q. infectoria subsp. infectoria were previously reported by Yilmaz et al. (Citation2008). In the comparison of two subspecies of Q. infectoria, very similar results were found with chromosome number and all metacentric chromosomes. Furthermore, it was observed that the chromosome sets of Q. infectoria subsp. boissieri and Q. infectoria subsp. infectoria were quite small and similar, 1.02–2.35 and 0.91–1.96 respectively.
| (2) | The Q. pubescens chromosome number was found to be 2n = 24 and all chromosomes were metacentric. Among the studied taxa Q. pubescens has the smallest chromosomes set, 1.01–2.01 μm, and haploid complement value, 16.89 μm. A2 had the lowest value (0.21) among all the studied taxa, while A1 had the second lowest value (0.19) after Q. vulcanica. Previously chromosome numbers and morphometric parameters of Q. pubescens were reported by D’emerico et al. (Citation2000). The chromosome number of this taxon was reported as 2n = 24 and the karyotype consists of 18 metacentric and six submetacentric chromosomes according to D’emerico et al. (Citation2000). | ||||
In this study, chromosome number was compatible with 2n = 24 but chromosome morphologies showed differences with all metacentric chromosomes. However, there were also differences in haploid complement value in comparison to the value reported (27.28) as a result of small chromosome set in this study. These differences can be caused by oaks living in different geographical regions and hybridization behaviours seen commonly between oak species living in mixed populations.
Q. pubescens has a wide distribution range, occupying almost all of western, central and southern Europe from Spain to Turkey. Frequent hybridization with other oaks and the fragmentation of its populations due to human impact are responsible for the remarkable variability. There are many hybrids of Q. pubescens, especially with Q. infectoria, Q. petraea and Q. macranthera subsp. syspirensis in Turkey (Yaltirik Citation1984).
| (3) | The chromosome number of Q. robur from the Uşak location was found compatible with previous reports, as 2n = 24 (D’emerico et al. Citation2000). Detailed chromosome analysis showed that all chromosomes were metacentric. This taxon has differences in chromosome lengths and haploid complement compared to the other studied taxa (Table ). The haploid complement value of Q. robur was found to be 31.78 and chromosome lengths ranged from 1.75 to 3.92 μm. Among the studied taxa Q. robur has the highest A1 value (0.22). | ||||
| (4) | Q. vulcanica is an endemic taxon from Turkey. The total chromosome number was observed as 2n = 24 with all metacentric chromosomes. The haploid complement has the second highest value with 22.63 μm and chromosomal lengths ranged from 1.25 to 3.13 μm. Among the studied taxa Q. vulcanica has the lowest A1 value (0.18) and the highest A2 value (0.28) (Table ). | ||||
Q. vulcanica has a restricted distribution in Isparta/Eğirdir and Afyon/Sultan Mountains in Turkey. As a result, the taxon is not well known and this is the first report of chromosome number and detailed karyotype of Q. vulcanica.
Generally most oak species examined until now have a diploid chromosome number with 2n = 24 (Mehra et al. Citation1972; Ohri and Ahuja Citation1990; D’emerico et al. Citation1995, Citation2000; Zoldo et al. Citation1998; Kurokawa and Yonezawa Citation2004; Yilmaz et al. Citation2008, Citation2011). In this study, the chromosome numbers of all analysed species were found to be 2n = 24 and compatible with previous reports. Consequently, the results of this study and other reports on the genus Quercus support that the basic chromosome number of the genus is n = 12. However, the presence of different chromosome number among oak species is reported by Zoldo et al. (Citation1998) and Chokchaichamnankit et al. (Citation2008). Zoldo et al. (Citation1998) state that chromosome number of Q. petraea may be 2n = 24 + 1,2,3. Chokchaichamnankit et al. (Citation2008) report that one tree belonging to Q. lenticellatus had 2n = 14.
Oaks, which dominate most forests of Turkey, and have a wide geographical distribution, are represented by 18 species in Turkey (Yaltirik Citation1984). Comparison of the results of this karyological study on Turkish oaks with previous reports shows that the chromosome number of the genus Quercus is stable with 2n = 24 (Yilmaz et al. Citation2011).
Although the findings of this study showed parallelism between oak taxa in Turkey, clear separations were detected between Turkish oaks and European oaks in many chromosomal characters like chromosomal length, haploid complement, intrachromosomal asymmetry (A1) and interchromosomal asymmetry (A2).
The basic reason for the similarity of the morphometric parameters obtained from the chromosomes of the species studied may be caused by the genetic similarity and gene flow between species belonging to the same section (section Quercus). However, study results together with previous reports on Quercus in Turkey showed less parametric values than Quercus species studied by D’emerico et al. (Citation1995). This situation may be a reason for extensive hybridization behaviours because of weak reproductive barriers between mixed oak populations in different geographical regions (Bacilieri et al. Citation1996; Manos et al. Citation1999; Borazan and Babaç Citation2003).
The genus is very important economically in Turkey and has been used for many purposes, such as fuel wood, furniture, tannins for leather, foods for animals. This not only increases taxonomic problems, but also makes it difficult to understand the relationships between oaks. For this reason, new management strategies for oak habitats should be produced to get the best result.
Among the studied taxa, the endemic species Q. vulcanica exhibited the highest variation in asymmetric index A1 and A2. This species is naturally distributed from 1200–2000 m altitude in restricted areas such as Kütahya-Türkmen Mountains, Konya-Sultan Mountains and Isparta-Eğirdir (Yukari Gökdere village). This high variation could be caused by the geographical distribution in this restricted area and more isolated habitats compared with other oak species. Quercus vulcanica has been faced with the threat of extinction because of over-exploitation for wooden home appliances, veneer and furniture. To protect this valuable resource 1300.5 hectares area near the Eğirdir Yukari Gökdere village was declared as a Nature Reserve Area for this endemic species.
This study of one endemic and three common Quercus taxa has indicated karyotypic diversification. In particular, the study results contribute to understanding the relations among Turkish oaks and European oaks. Another important contribution is the first report of Q. vulcanica. However, the study results demonstrate that comprehensive studies including karyotypes of many oak species are needed to understand oak taxonomy.
Disclosure statement
No potential conflict of interest was reported by the author.
Acknowledgements
The author would like to thank Uşak University Directorate of Scientific Research Projects (BAP) for providing financial support and also special thanks to Mustafa Kemal Pişmiş, Ayhan Yilmaz and İbrahim Öztürkmen for helping to collect plant material. Finally thanks to Ahmet Kahraman for his contribution to this manuscript.
References
- Bacilieri R, Ducousso A, Petit RJ, Kremer A. 1996. Mating system and asymmetric hybridization in mixed stand of European oaks. Evolution. 50:900–908.10.2307/2410861
- Borazan A, Babaç MT. 2003. Morphometric leaf variation in oaks (Quercus) of Bolu. Turkey Ann Bot Fenn. 40:233–242.
- Borgardt SJ, Pigg Kathleen B. 1999. Anatomical and developmental study of petrified Quercus (Fagaceae) fruits from the middle Miocene, Yakima Canyon, Washington. USA Am J Bot. 86(3):307–325.
- Chokchaichamnankit P, Anamthawat-Jonsson K, Chulalaksananukul W. 2008. Chromosomal mapping of 18S-25S and 5S ribosomal genes on 15 species of fagaceae from Northern Thailand. Silvae Genet. 57(1):5–13.
- D’emerico S, Bianco P, Medagli P, Schirone B. 1995. Karyotype Analysis in Quercus ssp. (Fagaceae). Silvae Genet. 44:66–70.
- D’emerico S, Paciolla C, Tommasi F. 2000. Contribution to the karyomorphology of some species of the genus Quercus. Silvae Genet. 49(6):243–245.
- Denk T, Grimm GW. 2010. The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon. 59(2):351–366.
- Dogan Y, Baslar S, Kanisanli M. 2000. Bati Anadolu’da Yayiliş Gösteren Quercus ithaburensis Decne supsp. macrolepsis (Kotschy) Hedge Et Yalt. (Fagaceae) (Palamut Meşesi) Üzerinde Bir Araştirma [An investigation on Quercus ithaburensis Decne supsp. macrolepsis (Kotschy) Hedge Et Yalt. (Fagaceae) (Palamut Meşesi) distribution in Western Anatolia]. Ekoloji Çevre Dergisi. 9(35): 22–25.10.5053/ekoloji
- Govaerts R, Frodin DG. 1998. World checklist and bibliography of Fagales (Betulaceae, Corylaceae, Fagaceae and Ticodenraceae). Great Britain: Royal Botanic Gardens, Kew.
- Kremer A, Petit RJ. 1993. Gene diversity in natural populations of oak species. Annales des sciences forestieres. 50(Supplement 1):186s–202s.10.1051/forest:19930717
- Kurokawa Y, Yonezawa Y. 2004. Karyotype analysis of fifteen species of Quercus L. (Fagaceae) in Japan. Chromosome Sci. 8(4):209.
- Levan A, Fredga K, Sandbdberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Manos PS, Doyle JJ, Nixon KC. 1999. Phylogeny, biogeography, and processes of molecular differentiation in Quercus Subgenus Quercus (Fagaceae). Mol Phylogenet Evol. 12:333–349.10.1006/mpev.1999.0614
- Maryam A, Fatima R, Abbas S. 2012. Genetic variation among Iranian oaks (Quercus ssp.) using random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol. 11(45):10291–10296.
- Mehra PN, Hans AS, Sareen TS. 1972. Cytomorphology of Himalayan Fagaceae. Silvae Genet. 21(3–4):102–109.
- Nixon KC. 2002. The Oak (Quercus) biodiversity of California and adjacent regions. USDA forest service gen. Tech Rep. PSW-GTR-184. San Diego, California.
- Ohri D, Ahuja MR. 1990. Giemsa C-banded Karyotype in Quercus L. (Oak). Silvae Genet. 39:216–219.
- Olfat AM, Pourtahmasi K. 2010. Anatomical characters in three Oaks species (Q. libani, Q. brantii and Q. infectoria) from Iranian Zagros Mountains. Aust J Basic Appl Sci. 4(8):3230–3237.
- Ribeiro T, Loureiro J, Santos C, Morais-Cecílio L. 2011. Evolution of rDNA FISH patterns in the Fagaceae. Tree Genet Genomes. 7(6):1113–1122.10.1007/s11295-011-0399-x
- Simeone MC, Piredda R, Papini A, Vessella F, Schirone B. 2013. Application of plastid and nuclear markers to DNA barcoding of Euro-Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. Bot J Linn Soc. 172(4):478–499.10.1111/boj.2013.172.issue-4
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold.
- Yaltirik F. 1984. Türkiye meşeleri teşhis kilavuzu [Diagnostic guide of Turkish oaks]. İstanbul: Yenilik Basimevi.
- Yilmaz A, Uslu E, Babaç MT. 2008. Karyological studies on four Quercus L. Species in Turkey Caryologia. 61(4):397–401.
- Yilmaz A, Uslu E, Babaç MT. 2011. Cytogenetic studies on Quercus L. (Fagaceae) species belonging to Ilex and Cerris sections in Turkey. Caryologia. 64(3):297–301.
- Zarco CR. 1986. A new method for estimating Karyotype asymmetry. Taxon. 35(3):526–530.10.2307/1221906
- Zoldos V, Papes D, Brown SC, Panaud O, Siljak-Yakovlev S. 1998. Genome size and base composition of seven Quercus species: inter- and intra- population variation. Genome. 41(2):162–168.10.1139/gen-41-2-162
