Abstract
The karyotype asymmetry/symmetry of a taxon (eight populations) from different geographic sites of Zhumeria genus is presented. The ploidy level between and within populations was 4x. According to intra-chromosomal asymmetry, population Z2 had the most symmetrical karyotype and population Z7 had the most asymmetrical evolutionary karyotype among the populations. In terms of the Stebbins system, the karyotype of populations can be classified in 1A, 2A and 2B classes. The results of analysis of variance revealed significant differences between the populations based on nine karyotypic traits (p < 0.05 and p < 0.01). Cluster analysis showed that the populations of Zhumeria majdae have been grouped in separate clusters. The results seemed to provide enough genetic evidence to identify different populations and useful data to clarify the interspecific relationships. Detailed karyotype analysis allows us to group the different populations and to postulate relationships among them.
Introduction
Zhumeria is a genus of flowering plant in the Lamiaceae family, first described in 1967. It contains just one known species, Zhumeria majdae Rech. f. & Wendelbo, endemic to Iran, known locally by the name of mohrekhosh, and it has a limited geographical range, very sharp slope of the mountains at 520–1450 m altitude in Bandar-Abbas (Hormozgan province) in the south of Iran (Rechinger and Wendelbo Citation1967; Rechinger Citation1982).
Zhumeria is unusual within the broader Salvia clade in that, in addition to the two fertile stamens, two large staminodes are easily identified in the corolla, and it is monotypic genus of Lamiaceae (Bokhari and Hedge Citation1976). It is a perennial fragment shrub, evergreen and without any spine.
Research by Walker and Sytsma (Citation2007) showed that Z. majdae could be placed as sister to one of the two groups in clade III of Salvia genus. They informally recognize within the larger tribe Mentheae a lineage that would correspond to a sub tribe consisting of the genera Salvia, Dorystaechas, Meriandra, Zhumeria, Perovskia, Rosmarinus, Lepechinia and Melissa (Walker and Sytsma Citation2007; Will and Claßen-Bockhoff Citation2014).
The leaves have been used for many years as a curative for stomach aches, as an antiseptic, as a carminative especially in infants and for treatment of painful menstruation (Safa et al. Citation2013).
The extract of aerial parts of the plant has shown anti-nociceptive effects in mice and rats (Hosseinzadeh et al. Citation2002). The ethanol extract of Z. majdae has shown potent anti-leishmanial and anti-plasmodial activity in vitro (Moein et al. Citation2008). Antibacterial activity of the essential oil of Z. majdae has been previously reported only against two bacterial strains of S. aureus and E. coli (Soltanipoor Citation2007). The aim of the present work was to study mitosis of eight populations of Z. majdae in all points of distribution in Iran, trying to reveal the chromosome numbers and basic cytogenetic information.
Materials and methods
In this study, we used root tip meristems from seedlings obtained by the germination of ripe seeds collected from eight natural populations on wet filter paper in Petri dishes at 22°C. The studied populations are listed in (Table ).
Table 1. Ecological characteristics of Zhumeria majdae localities.
Vouchers are deposited in RIFR gene bank (Research Institute of Forest and Rangelands in Iran). Root tip meristems obtained from seedlings were pretreated with 0.7% saturated α-bromo naphthalene at 4°C for 3.5 h, fixed in 10% formaldehyde and 1% chromic acid (1:1) for at least 15 h at room temperature, then root tips were rinsed for 2 h in distilled water. Hydrolysis was carried out with 1 N NaOH at 60°C for 10 min, dyed with hematoxylin-iron for 4 h and squashed in a droplet of 45% acetic acid and lactic acid (10:1) (Wittmann Citation1965; Hesamzadeh Hejazi and Rasouli Citation2006; Hesamzadeh Hejazi and Ziaei Nasab Citation2010). The preparations were observed with an optical microscope (BH2 Olympus supplemented digital color video camera) at a magnification of 2000 × . The best metaphase plates were selected and measured by Micromeasure 3.3 software (Reeves and Tear Citation2000). In each mitotic metaphase (at least 10 plates) the arm length of each chromosome was measured. The parameters such as long arm (LA), short arm (SA), total length (TL), relative length percentage (RL %), arm ratio (AR), centromeric index (CI), and value of relative chromatin (VRC) were estimated in each metaphase plate to characterize the karyotypes numerically (Martinoli and Ogliotti Citation1970; Hesamzadeh Hejazi and Ziaei Nasab Citation2009, Citation2010). Karyotype asymmetry was estimated by four different methods namely, total form percentage (TF %) (Huziwara Citation1962); difference of relative length (DRL); intra-chromosomal asymmetry index (A1) and inter-chromosomal asymmetry index (A2) (Romero Zarco Citation1986). Both indices (A1 and A2) are independent to chromosome number and size. Dispersion index (DI) (Lavania and Srivastava Citation1992) and also karyotypic evolution by using the symmetry classes of Stebbins (SC) (Stebbins Citation1971) has been determined.
The karyotype formula was determined according to classification of Levan (Levan et al. Citation1964). For each population, karyograms were adjusted based on length of chromosome size (arranged large to small). In order to determine the variation between populations, one-way balanced ANOVA was performed on normal data and parameter means were compared by Duncan’s test. The principal components analysis (PCA) was performed to evaluate the contribution of each karyotypic parameter to the ordination of species. Clustering was performed using Ward’s cluster analysis method after calculation of Cophenetic correlation coefficient (r) to examine karyotype similarity among populations. FCM analysis was used to reconfirm the ploidy level of various populations. To determine the standard peak of diploid cells (2C DNA) in FCM analysis, at least 10 leaves were collected from diploid salvia reuteriana (2n = 2x = 20) plants grown in the greenhouse.
Results
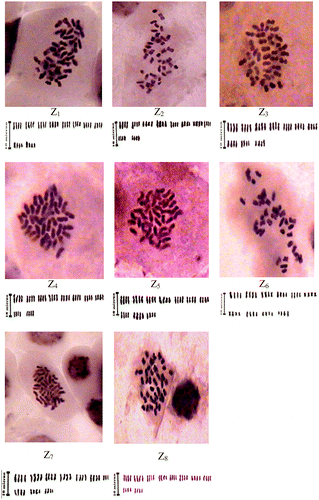
This study reveals a detailed picture of the chromosome features in Z. majdae. Pictures of the mitotic metaphases and karyograms of the populations are presented in Figure . The somatic chromosome numbers (2n), ploidy levels, ranges of chromosome length, symmetry index percentage, intra and inter-asymmetry indices, difference of range relative length, total form percentage, symmetry classes, total karyotype length and karyotype formula of the taxa and populations investigated are summarized in Table . The somatic chromosome number and details of the karyotypes of the studied populations, revealed that Z. majdae populations possessed one ploidy level (2n = 4x = 40).
Table 2. Somatic chromosome number (2n), ploidy levels, ranges of chromosome length, arm ratio (AR), Romero Zarco asymmetry indices (A1, A2), dispersion index (DI), difference of range relative length (DRL), total form percentage (TF%), Stebbins symmetry classes (SC), total karyotype length (TKL) and karyotype formula (KF) (m: metacentric, sm: submetacentric).
Total karyotype length, roughly indicative of the DNA content, ranges from 46.95 to 58.45 μm in all populations. Also size of the chromosomes among the populations varied from 1.32 μm in population Z8 to 4.05 μm in population Z4. Most of the chromosomes were mainly “m” type chromosomes (centromeres at median region). Another type of chromosome was “sm” type chromosomes (centromeres near median region).
Among the studied populations, the highest TF% value (43.21) was estimated in the Z2 population and the lowest TF% value (36.72) was estimated in the Z7 population, the lowest of TF% value is one of the main reasons to make karyotype asymmetric in Z7 population. In view of the fact that lower DRL value indicated more karyotype symmetry, the Z2 and Z8 populations, with DRL 2.60 and 3.59 values, respectively, had the most symmetric and asymmetric karyotypes.
Intrachromosomal asymmetry index (A1) showed sharp differences between the chromosome arms in the different populations. In general, based on intrachromosomal asymmetry (A1 and TF %), population Z7 had the most asymmetric karyotype and population Z2 had the most symmetrical karyotype in all of the populations.
In terms of the Stebbins system, the karyotype of populations seizes 1A, 2A and 2B classes which are considered mainly primitive classes in this system. In this study, Z. majdae (Z6) had a higher DI value, which is associated with an enhanced order of karyotypic specialization. Z. majdae (Z6) had the highest A2 values; therefore its karyotype was more asymmetric than the other populations in the same symmetry classes (Tables and ).
To analyze the variability of the karyotypes among populations, length of chromosome, long and short arms of chromosome, arm ratio values, difference of range relative length, total form percentage and asymmetry indexes (A1, A2) were compared by one-way analysis of variance based on completely randomized design (CRD). Also, the Duncan test was carried out to test differences between each pair of means. The results of variance analysis revealed significant differences between the populations based on seven karyotypic characteristics (p < 0.05 and p < 0.01) (Table ). The Duncan test applied to the chromosome morphometric traits showed a highly significant difference among all examined populations (Table ). So, mean values of chromosome total length varied from 2.35 μm in populations Z1 to 2.92 μm in populations Z4. The mean values of chromosome long arms varied from 1.52 μm in populations Z2, and Z6 to 2.27 μm in population Z1. Also the mean values of chromosome short arms was different from 0.93 μm in populations Z1 and Z8 to 1.15 μm in populations Z2 and Z4.
Table 3. The results of analysis of variance for karyotypic data based on CRD design.
Table 4. Mean of parameters of chromosomes analysis of Zhumeria majdae populations.
Using principal components analysis (PCA), the first two independent components accounted for about 84.55% of total variation. The first component emphasized short arm, arm ratio, total form percentage, intrachromosome asymmetry index, long arm and short arm percentage value which had the highest coefficients of Eigen vectors and were important characters for classification of populations with about 59.4% of total variation. Interchromosome asymmetry index was important trait in the second component (25.15%) (Table ).
Table 5. Eigenvectors from the first two principal components for seven karyotype parameters to classify eight populations of Zhumeria majdae.
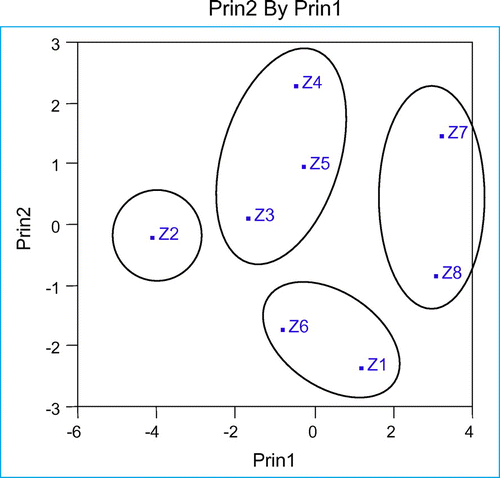
Grouping of studied populations was based on their karyotypic traits (Figure ). The results showed that populations of Z. majdae have been grouped in separate cluster.
Figure 3. Dendrogram of eight populations of Zhumeria majdae by analyzing seven karyotypic parameters using Ward’s cluster analysis method. Cophenetic correlation r = 0.88.
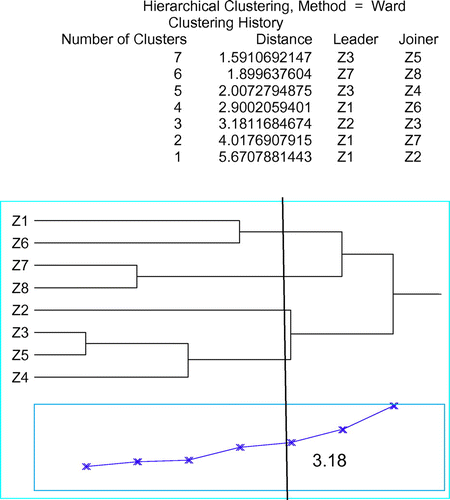
By cutting the dendrogram resulting from cluster analysis by Ward’s method with cophenetic correlation coefficient (r = 0.88) with a metric distance of 3.18, the populations were classified into four groups. The highest metric distance (5.67) was obtained between Z1 and Z2, which implies the least affinity between them. The lowest metric distance (1.59) was obtained between Z3 and Z5, which implies the least karyotypic difference between them (Figure ). The diagram of population dispersion, based on the first two components, showed that the populations separated in four groups, which completely fits with results obtained through the grouping analysis by Ward’s method (Figure ).
Population of Z2 is classified as a separate group. Two populations of Z7 and Z8, two populations of Z1 and Z6 and three populations of Z3, Z4, Z5 are grouped together in the same cluster.
The population of Z2 had the lowest value of A1 with 0.22 and the highest TF% with 43.21%. The populations of Z7 and Z8 had the highest values of A1 (0.28–0.31) and the lowest values of TF% (36.72–37.56). The populations of Z1 and Z6 were grouped together in the same cluster that had the highest value of A2, with 0.21. The populations of Z3, Z4 and Z5 were grouped together in the same cluster, although some of the factors were statistically significant (Table ). The ranges of A1, A2 and TF% values were 0.29–0.33, 0.136–0.155 and 39.36–41.12 respectively (Table and Figure ).
Discussion
In this study, chromosome numbers and detailed measurements of eight populations of the Zhumeria genus were determined for the first time in Iran. The karyotype concept has been used in characterizing and distinguishing chromosomes of different populations. Mitotic karyotype analyses are also helpful in studying evolutionary problems. Bokhari and Hedge (Citation1976) reported that one population of Z. majdae located in Hormozgan province in Iran has 2n = 40 or 42. Our study indicated that the chromosome numbers in different populations of Hormozgan and Fars province are tetraploid (2n = 4x = 40). To reconfirm the ploidy level, flow cytometric analysis was performed using a PA-1 (Partec, GmbH/CyFlow space-Munster) flow cytometer. All the studied populations fitted a 4x ploidy level in compare with salvia reuteriana as a standard plant (2n = 2x = 20). By using A1 and A2 parameters, we can determine the more asymmetric karyotype among the populations which have the similar Stebbins classes of symmetry. The populations which are classified as 1A group also showed the lowest value of A1 in the range of 0.22–0.29 and the highest value of TF%, ranging from 41.10 to 43.21. In order to refine the measure of karyotype asymmetry we used the DI, which has the potential to decipher even minor karyotypic variations. The DI index plays an important role in arranging the species within the same class of karyotype asymmetry in an advancing order of specialization by permitting further gradations, as depicted by species arrangement within sections. The results of variance analysis between the populations revealed significant differences based on karyotypic characteristics. This result indicated the occurrence of quantitative changes in chromosome size of the studied populations. A significant effect of chromosomal traits proved karyotypic variation between populations. This shows the importance of chromosome study to distinguish the state of evolution and affinity between different populations.
This study indicates that populations of a specific species in within itself will show variety. The important factors in separation of populations were inter and intra chromosome asymmetry factors (A1, A2 and TF% values). Thus, these studies could greatly help us in classification and taxonomic studies. The present study shows the change in chromosomal traits as one of the mechanism of inter and intra-species diversification in the Zhumeria genus, as well as the earlier cytological reports (Hesamzadeh Hejazi and Ziaei Nasab Citation2009; Javadi et al. Citation2009; Hesamzadeh Hejazi and Ziaei Nasab Citation2010; Hesamzadeh Hejazi Citation2011; Kalvandi et al. Citation2012; Ziaei Nasab et al. Citation2012; Salehi et al. Citation2014). In this research we found that there are differences in position of centromere and total length of chromosomes, while there are no morphological differences among populations in Zhumeria genus. So, karyotype study may show variations which do not appear in the morphology of the plant.
Investigation of different Zhumeria populations showed that they grow in locations with varying climate, height, soil, slope, vegetative type and geographical characteristics. For this reason some of the populations are separated or are grouped together.
The essential oils from eight populations studied in this paper also have been chemically investigated. The results showed that there are about 25 different volatile components in total. The amount of components varied between populations (data not shown). So it seems the different components of essential oils in different populations of Z. majdae are variable due to hybridization and geographical location.
The results of the molecular markers used among the eight populations also showed a high diversity as well as the amounts of components of essential oils (data not shown).
The variation of climate and soil in Iran provides a suitable field for plant variations. One of the clearly detectable genetic variations in Z. majdae is the structural changes of chromosomes. These genomic differences could be used for breeding purposes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by the biotechnology department of Research Institute of Forests and Rangelands (RIFR), Iran.
Acknowledgements
The authors are grateful to the Hormozgan Agricultural and Natural Resources Research Center for providing seedlings and seeds.
References
- Bokhari M, Hedge I. 1976. Zhumeria (Labiatae): anatomy, taxonomy and affinities. Iran J Bot. 1(1):1–10.
- Hesamzadeh Hejazi SM, Rasouli M. 2006. Cytogenetic study of some species of vetch genus (Vicia sp.) in Iran. Iran J Agric Sci. 37-1(2):213–225.
- Hesamzadeh Hejazi SM, Ziaei Nasab M. 2009. Cytogenetic study on several populations of diploid species of Onobrychis in natural gene bank of Iran. Iran J Rangelands For Plant Breed Genet Res. 16(2):158–171.
- Hesamzadeh Hejazi SM, Ziaei Nasab M. 2010. Cytotaxonomy of some Onobrychis (Fabaceae) species and populations in Iran. Caryologia. 63(1):18–31.
- Hesamzadeh Hejazi SM. 2011. Karyological study on three Cicer L. species (Fabaceae) in Iran. Asian J Cell Biol. 6(3):97–104.
- Hosseinzadeh H, Ramezani M, Fadishei M, Mahmoudi M. 2002. Anti-nociceptive, anti-inflammatory and acute toxicity effects of Zhumeria majdae extracts in mice and rats. Phytomedicine. 9(2):135–141. 10.1078/0944-7113-00097
- Huziwara Y. 1962. Karyotype analysis in some genera of compositae. VIII. Further studies on the chromosome of Aster. Am J Bot. 49(2):116–119. 10.2307/2439026
- Javadi H, Hesamzadeh Hejazi SM, Majnoon SHB. 2009. Karyotipic studies of three Thymus (Lamiaceae) species and populations in Iran. Caryologia. 62(4):316–325.
- Kalvandi R, Hesamzadeh Hejazi SM, Atri M, Mirza M, Jamzad Z, Safikhani K. 2012. Karyotype analysis among 10 populations of Thymus eriocalyx spesies in Iran. Ann. Biol Res. 3(8):3916–3925.
- Lavania UC, Srivastava S. 1992. A simple parameter of dispersion index that serves as an adjunct to karyotype asymmetry. J Biosci. 17(2):179–182. 10.1007/BF02703503
- Levan AK, Fredga K, Sandberg AA. 1964. Nomenclature for centromic position on chromosomes. Hereditas. 52(2):201–220.
- Martinoli G, Ogliotti P. 1970. Ricerchecitotassonomiche in Artemisia vulgaris L. ed Artemisia verlotorum Lamotte. G Bot Ital. 104(5): 373–387. 10.1080/11263507009426510
- Moein MR, Pawar S, Khan I, Tekwani L, Khan A. 2008. Antileishmanial, antiplasmodial and cytotoxic activities of 12,16-dideoxy aegyptinone B from Zhumeria majdae. Phytother Res. 22(3):283–285. 10.1002/(ISSN)1099-1573
- Rechinger K. 1982.Flora Iranica, Labiatae, Akademische Druke- u.Verlagsanstalt. Graz Austria., 150:479–480.
- Rechinger K, Wendelbo P. 1967. Zhumeria majdae. Nytt. Magazine Botanikk (Oslo) 14(1):39–43.
- Reeves A, Tear J. 2000. Micromeasure software. Colorado State University. Available from: http://www.colostate.edu/Depts/Biology/micromeasure.
- Romero Zarco C. 1986. A new method for estimating Karyotype asymmetry. Taxon. 35(3):526–530.
- Safa O, Soltanipoor MA, Rastegar S, Kazemi M, Nourbakhsh Dehkordi M, Ghannadi A. 2013. An ethnobotanical survey on Hormozgan province. Iran Avicenna J Phytomed. 3(1):64–81.
- Salehi M, Hesamzadeh Hejazi SM, Tabaei aghdaei SR. 2014. Cytogenetic studies of two Dracocephalum (Lamiaceae) species and populations in Iran. Int J Biosci. 4(9):100–108.
- Soltanipoor MA. 2007. Investigation of Ecological factors on abundant and distribution of Zhumeria majdae. Pajouhesh Sazandegi Mag. 76(3):54–61.
- Stebbins GL. 1971. Chromosomal evolution in higher plants. London: Edward Arnold Publisher Ltd; p. 216.
- Walker JB, Sytsma KJ. 2007. Staminal evolution in the genus Salvia (Lamiaceae): molecular phylogenetic evidence for multiple origins of the staminal lever. Ann Bot. 100(2):375–391. 10.1093/aob/mcl176
- Will M, Claßen-Bockhoff R. 2014. Why Africa matters – evolution of old world Salvia L. (Lamiaceae) in Africa. Ann Bot - London. 114(1):61–83. 10.1093/aob/mcu081
- Wittmann W. 1965. Aceto-iron-haematoxylin-chloral hydrate for chromosome staining. Stain Technol. 40(3):161–164. 10.3109/10520296509116398
- Ziaei Nasab M, Hesamzadeh Hejazi SM, Bihamta MA, Mirza M, Naderi-Shahab MA. 2012. Assessment of Karyotypical variation among 16 populations of Thymus daenensis Cleak and Thymus kotchyanus Boiss. species in Iran. Afr J Biotechnol. 11(5):1028–1036.