Abstract
The influence of biotic, abiotic and climatic changes on crop production is becoming increasingly evident. Gaps in demand and supply of lentils, one of the few protein-rich crops in India, are gradually increasing. Of seven estimated species of lentil, six are wild and the other, Lens culinaris, is the only species under cultivation, with a large number of cultivars. Chromosome analysis is beneficial to breeders and genome researchers in crop improvement programmes. However, a chromosomal database of this important crop is not available in India. The present paper has described a detailed chromosome analysis of 21 certified Indian cultivars of L. culinaris using enzymatic maceration and air drying (EMA) based Giemsa staining methods for the first time. Uniform chromosomal preparations have resulted in variation in average chromosome size (4.94–9.8 μm), total chromatin length (69.18–137.24 μm), and satellite bearing chromosome number (third, fourth and fifth) among the cultivars. Though our results revealed similar karyotypic formulae and symmetrical karyotype, a scatter diagram of intra-chromosomal asymmetry index (A1) versus inter-chromosomal asymmetry index (A2) groups them into five distinct clusters. Such information may be helpful for conservation of genetic diversity and for future lentil breeding programmes.
Keywords:
Introduction
The only worldwide cultivated lentil species, Lens culinaris Medik. (Fabaceae), is popularly known as “poor man’s meat” due to its high protein content (Jha et al. Citation2015). The genus comprises seven taxa, of which six are reported as wild species (van Oss et al. Citation1997). Lentil is gradually becoming a costly pulse in the Indian national market primarily due to its high domestic consumption. India is the second highest producer of lentil and different cultivars are encouraged for large-scale cultivation. Increase of cultivation acreage as well as improvement of the crop against biotic and abiotic stresses are important. Crop improvement, conservation and utilization of plant genetic resources have benefited from chromosomal studies of the available germplasms (Mishra et al. Citation2007). Cultivated and wild germplasms of lentil are reported to have 2n = 2x = 14 chromosomes (Mehra et al. Citation1986; Jha et al. Citation2015; Jha and Halder Citation2016). Analysis of genetic constitution is an important parameter that aids breeders and researchers in their crop improvement programmes. Many researchers from different countries have carried out conventional and molecular cytogenetics, genomic DNA analysis and next generation sequencing in lentil species and cultivars (Ladizinsky Citation1979; Galasso Citation2003). Although India is considered an important lentil producing country with a large number of cultivated germplasms, the country does not have a chromosomal database. One may consider traditional karyotype analysis an obsolete method in the post genomic era, but chromosomes represent genomic configuration, and basic karyotypic information such as chromosome number, size and organization, and position of chromosomal landmarks are prerequisites for correct data interpretation in next-generation sequencing (Weiss-Schneeweiss and Schneeweiss Citation2013). Moreover, conservation of chromosomal information may help genome researchers in formulating new crop improvement strategies.
Chromosomal analysis of lentil with the conventional squash method was started in India primarily on cultivated species more than six decades ago (Bhattacharjee Citation1953; Sharma and Mukhopadhyay Citation1963; Sinha and Acharia Citation1972; Naithani and Sarbhoy Citation1973; Lavania and Lavania Citation1983; Nandanwar and Narkhede Citation1991). However, systematic chromosomal analysis on this ancient crop is still lacking. We have collected more than 35 certified germplasms of cultivated and wild species of lentil from the Indian Institute of Pulses, Kanpur, India. For the first time we have applied enzymatic maceration and air drying (EMA) methods for chromosome preparation in Indian lentils, to produce a chromosomal database. Our first report documented detailed karyotypic analysis of 14 Indian germplasms (Jha et al. Citation2015). Our second report highlighted comparative karyomorphology of one cultivated and four wild species and documented new chromosomal landmarks in two wild species (Jha and Halder Citation2016). The present research work has analysed 21 lentil germplasms belonging to L. culinaris and prepared karyotypes, comparative idiograms and scatter diagrams, with the aim of conservation of genetic diversity as well as for lentil breeders.
Materials and methods
Plant material
In the present study, 21 Indian cultivars of Lens culinaris were considered for detailed cytological study. One cultivar was collected from Uttar Pradesh, and the other 20 samples were obtained from the Indian Institute of Pulses, Kanpur, India. Collection details of the accessions along with their seed germination efficiency have been included in Table . A minimum of 10–15 seeds of each cultivar were immersed in water for 10–12 h, then placed on moist filter paper and kept in the dark for 48 h for germination.
Table 1. Germination efficiency of Lens culinaris cultivars used for EMA based chromosome analysis.
Chromosome preparation and karyomorphometric analysis
At least 10 healthy root tips collected from germinating seeds of each cultivar were pre-treated with saturated para-dichlorobenzene (pDB) at 14–16 °C for 4–5 h. They were fixed in 1:3 acetic-methanol overnight and finally stored at –20 °C for chromosome analysis. The EMA method was carried out for each cultivar following our earlier published protocol (Jha et al. Citation2015). Completely air dried slides were stained with 2% Giemsa solution (Merck, Darmstadt, Germany) in 1:15 phosphate buffer solution for 10–15 min at room temperature. After 4–5 times washing with doubled distilled water, the slides were air dried and observed under a light microscope (Axio Lab A1, Carl Zeiss, Jena, Germany). A minimum of 30 well scattered metaphase plates for each cultivar were examined under a Carl Zeiss Axio Lab A1 microscope using Axiovision L. E4 software (Carl Zeiss, Jena, Germany, https://www.zeiss.com/microscopy/int/products/microscope-software/axiovision.html).
For karyomorphometric analysis, different karyological parameters were assessed: length of long arm (l) and short arm (s), absolute chromosome length (CL), relative chromosome length (RL), average chromosome length (ACL) and total diploid chromatin length (TCL) were used. The arm ratio (r = l/s) was calculated to determine the centromeric position (Levan et al. Citation1964). Chromosome morphometric data were used to prepare ideogram and karyotype formulae of each of the cultivars of L. culinaris. Comparative karyometric analysis was performed among all the collected samples using intra-chromosomal asymmetry index (A1) and inter-chromosomal asymmetry index (A2) (Zarco Citation1986).
Statistical analysis
Means and standard deviations were analysed for all measured parameters and one-way analysis of variance (ANOVA) was performed to detect significant differences (p ≤ 0.05) in the mean (Sokal and Rohlf Citation1995). Duncan’s multiple range test (DMRT) was used for post hoc analyses using SPSS v 16.0 statistical package (SPSS Inc. (IBM), Chicago, http://www.spss.com.hk/corpinfo/index.htm). To study the relationships among the collected cultivars of L. culinaris, a scatter diagram of A1 versus A2 was also drawn, as described by Paszko (Citation2006).
Results
The present study reports on seed germination efficiency and karyomorphometry in 21 cultivars of L. culinaris. Differences in seed germination efficiency (70–95%) have been noted within all the studied cultivars (Table ).
Karyomorphometric analysis
For karyomorphometric analysis, an EMA based Giemsa staining method was conducted for all the collected Indian germplasms of L. culinaris. A minimum of 10 root tips per cultivar and about 30 countable metaphase plates were studied to determine the diploid chromosome number of the species. The present somatic chromosomal analysis revealed that all the cultivars of lentil (L. culinaris) possessed 2n = 2x = 14 chromosomes (Figures ; Table ). The presence of some monosomic cells (2n – 1) with 2n = 13 chromosomes was also noticed in three studied cultivars: DPL-15, DPL-62 and EC-78410. A significant variation in the chromosome size range was observed among the studied cultivars (Table ). The highest ACL (9.80 ± 0.01 μm) was found in EC-78475, while it was lowest (4.94 ± 0.09 μm) in EC-78461 (Table ). Interestingly, the total diploid chromatin length (TCL) ranging from 137.24 ± 0.17 μm to 69.18 ± 1.22 μm was found to be correlated with the ACL among the studied cultivars of lentil (Table ). All 21 studied cultivars of L. culinaris showed a common haploid karyotype formula, i.e.3 m+1 m.Sat+2Sm+1St, where the majority of chromosomes were found to have nearly median constrictions and were large in size (11.63 ± 0.12 μm to 5.87 ± 0.13 μm; Table ). The detailed karyomorphological analysis of the cultivars revealed the presence of a single satellited pair in their chromosome complements (Figures ). However, the ordering of satellite bearing pairs was found to be cultivar specific (Table ). In the present study, it was observed that out of 21, 15 cultivars bear satellite in chromosome number 4. On the other hand, four cultivars, IPL-406, HUL-57, EC-78475 and EC-267569-A, have their satellite in chromosome number 3 and in the case of EC-78542-A and EC-70394 it was the second and fifth chromosome pair respectively (Table ). The present comparative chromosome analysis documented several intraspecific karyomorphological variations in detail. Very low values of intra-chromosomal asymmetry index (A1) (≤ 0.04) and inter-chromosomal asymmetry index (A2) (< 0.07) indicate a more or less symmetrical karyotype in each of the studied cultivar (Table ).
Figure 1. Giemsa stained somatic metaphase plates of different cultivars of Lens culinaris. (a) DPL-15; (b) DPL-62; (c) IPL-406; (d) IPL-526; (e) HUL-57; (f) EC-70306; (g) EC-70394; (h) EC-70404. Arrows indicate chromosomes with two constrictions. Bar = 5 μm.
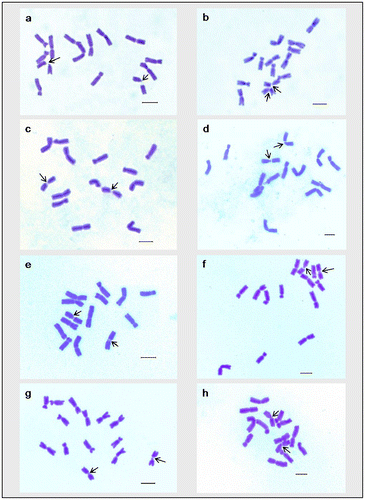
Figure 2. Giemsa stained somatic metaphase plates of different cultivars of Lens culinaris. (a) EC-78410; (b) EC-78451-A; (c) EC-78452; (d) EC-78475; (e) EC-78476; (f) EC-78542-A; (g) EC-223188; (h) EC-255491. Arrows indicate chromosomes with two constrictions. Bar = 5 μm.
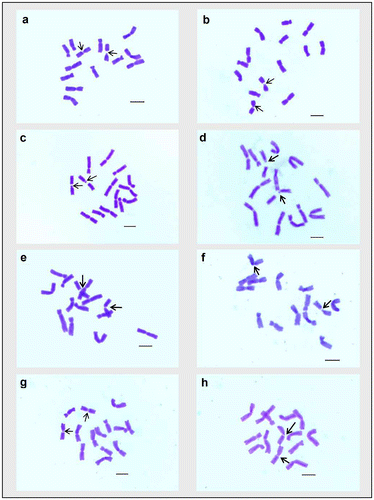
Figure 3. Giemsa stained somatic metaphase plates of different cultivars of Lens culinaris. (a) EC-267590; (b) Macro-Uttar Pradesh; (c) EC-267569-A; (d) EC-78461; (e) EC-78473. Arrows indicate chromosomes with two constrictions. Bar = 5 μm.
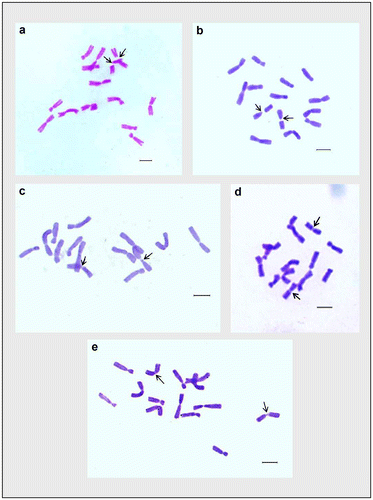
Figure 4. Hand drawings of different cultivars of Lens culinaris showing intercalary sat chromosome. (a) DPL-15; (b) DPL-62; (c) IPL-406; (d) IPL-526; (e) HUL-57; (f) EC-70306; (g) EC-70394; (h) EC-70404. Arrows indicate chromosomes with secondary constrictions. Bar = 5 μm.
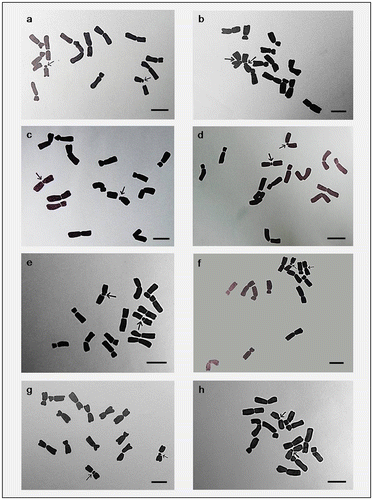
Figure 5. Hand drawings of different cultivars of Lens culinaris showing intercalary sat chromosome. (a) EC-78410; (b) EC-78451-A; (c) EC-78452; (d) EC-78475; (e) EC-78476; (f) EC-78542-A; (g) EC-223188; (h) EC-255491. Arrows indicate chromosomes with secondary constrictions. Bar = 5 μm.
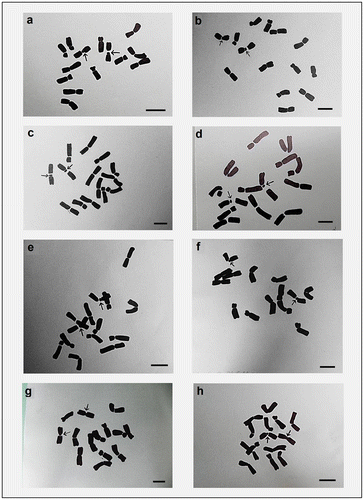
Figure 6. Hand drawings of different cultivars of Lens culinaris showing intercalary sat chromosome. (a) EC-267590; (b) Macro-Uttar Pradesh; (c) EC-267569-A; (d) EC-78461; (e) EC-78473. Arrows indicate chromosomes with secondary constrictions. Bar = 5 μm.
Table 2. Karyomorphometric data of 21 Indian cultivars of Lens culinaris.*
Figure 7. Comparative ideograms of different cultivars of Lens culinaris (2n = 14). (a) DPL-15; (b) DPL-62; (c) IPL-406; (d) IPL-526; (e) HUL-57; (f) EC-70306; (g) EC-70394; (h) EC-70404. Bar = 1 μm.
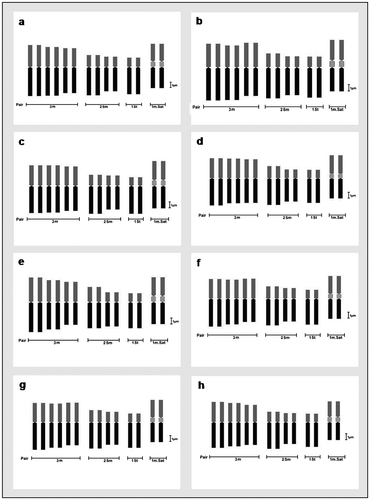
Figure 8. Comparative ideograms of different cultivars of Lens culinaris (2n = 14). (a) EC-78410; (b) EC-78451-A; (c) EC-78452; (d) EC-78475; (e) EC-78476; (f) EC-78542-A; (g) EC-223188; (h) EC-255491. Bar = 1 μm.
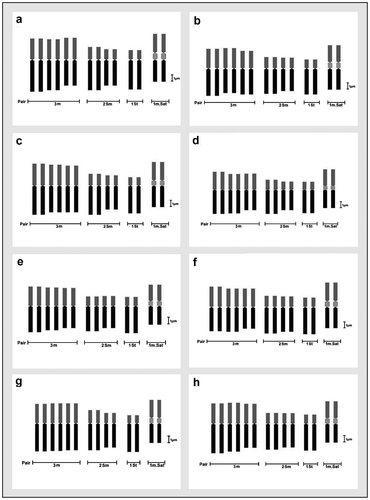
Figure 9. Comparative ideograms of different cultivars of Lens culinaris (2n = 14). (a) EC-267590; (b) Macro-Uttar Pradesh; (c) EC-267569-A; (d) EC-78461; (e) EC-78473. Bar = 1 μm.
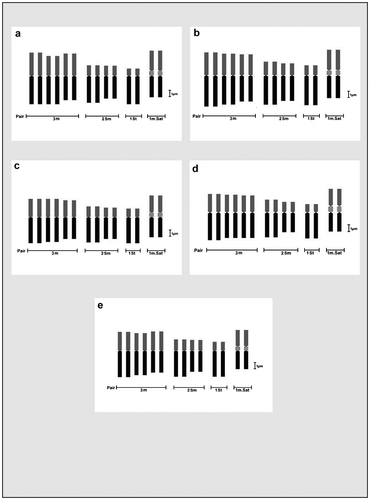
Table 3. Intra-chromosomal and Inter-chromosomal asymmetry index of 21 Indian cultivars of Lens culinaris.*
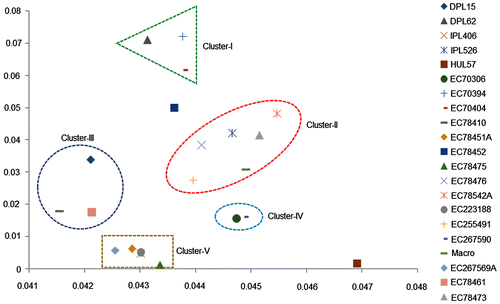
Although all the studied Indian cultivars of L. culinaris showed symmetrical karyomorphology, the scatter diagram of A1 versus A2 revealed the formation of cultivar specific clusters (Figure ). Formation of five distinct clusters (I to V) clearly demonstrated the intraspecific relationship among the studied 21 samples at cultivar level. Clusters I and III were composed of three cultivars each, while cluster II was the largest with six cultivars. Cluster IV included only two cultivars, whereas cluster V had five cultivars (Figure ). Interestingly, it was observed that one cultivar, EC-78452, placed equidistantly from clusters I and II, indicating its intermediate position between the members of clusters I and II in the scatter plot (Figure ). The present A1 versus A2 scatter diagram also showed that HUL-57 was positioned far away from all the five clusters (I to V), demonstrating its distant position from the rest of the cultivars.
Discussion
The results obtained from the present EMA based cytological analysis offer several insights into the chromosomal characterization of 21 different cultivars of this species. The combination of enzymatic maceration and air drying methods is a very useful technique to analyse chromosome morphology, constrictions and types of chromosomes in detail (Fukui Citation1996; Jha and Saha Citation2017). This method allows us to obtain uniformly spread chromosomes against a cytoplasm free background (Fukui Citation1996). The presence of diploid chromosome numbers (2n = 2x = 14) in all the studied cultivars of L. culinaris has corroborated the earlier reports (Mehra et al. Citation1986; Jha et al. Citation2015; Jha and Halder Citation2016). It is interesting to note that although a significant variation in chromosome size has been scored among the lentil cultivars, the haploid karyotype formulae was found to be common among them (Table ). Ladizinsky (Citation1979) reported that L. culinaris has three groups of chromosomes – submetacentric, metacentric including satellite and acrocentric – while in the present analysis it has been noted that all studied accessions of this species have four pairs of nearly metacentric chromosomes, where one pair includes satellite chromosomes, two pairs of submetacentric and one pair of subterminal chromosomes. The prevalence of a single satellited pair among the studied cultivars with a little variation in the ordering of satellite bearing pairs (Table ; Figures ) was in accordance with earlier reports (Mehra et al. Citation1986; Jha et al. Citation2015; Jha and Halder Citation2016). Moreover, in the present study, all the studied cultivars of L. culinaris with symmetrical karyotype explicitly formed distinct clusters based on their intra-chromosomal asymmetry index (A1) and inter-chromosomal asymmetry index (A2). Such clustering clearly demonstrated the intraspecific relationship within the Indian L. culinaris at cultivar level (Figure ).
In summary, this study provides comprehensive knowledge on karyotypes of 21 Indian cultivars of L. culinaris; this could be utilized as important background information for future lentil breeding programmes.
Disclosure statement
The authors declare that they have no conflict of interest.
Funding
This work was supported by University Grants Commission [F.No. 43-113/2014(SR)].
Acknowledgements
TBJ acknowledges UGC for awarding the project and Director and Dr J. Kumar of Indian Institute of Pulses, Kanpur, India for providing germplasms along with the Principal, Maulana Azad College, Kolkata for providing facilities.
References
- Bhattacharjee SK. 1953. Cytogenetics of Lens esculenta Moench. Caryologia. 5(2):159–166. doi:10.1080/00087114.1953.10797436
- Fukui K. 1996. Plant chromosomes at mitosis. In: Fukui K, Nakayama S, editors. Plant chromosomes: laboratory methods. Boca Raton (FL): CRC Press; p. 1–18.
- Galasso I. 2003. Distribution of highly repeated DNA sequences in species of the genus Lens Miller. Genome. 46(6):1118–1124. doi:10.1139/g03-077
- Jha TB, Halder M. 2016. Searching chromosomal landmarks in Indian Lentils through EMA-based Giemsa staining method. Protoplasma. 253(5):1223–1231. doi:10.1007/s00709-015-0873-7
- Jha TB, Mahanti A, Ghorai A. 2015. Karyotype analysis of Indian lentils through EMA-based Giemsa staining. Caryologia. 68(4):280–288. doi:10.1080/00087114.2015.1109921
- Jha TB, Saha PS. 2017. Characterization of some Indian Himalayan Capsicums through floral morphology and EMA-based chromosome analysis. Protoplasma. 254(2):921–933. doi:10.1007/s00709-016-1001-z
- Ladizinsky G. 1979. The origin of lentil and its wild gene pool. Euphytica. 28(1):179–187. doi:10.1007/BF00029189
- Lavania UC, Lavania S. 1983. Karyotype studies in Indian pulses. Genet Agrar. 37(1):299–308.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52(2):201–220.
- Mehra RC, Butler MG, Beckman T. 1986. N banding and karyotype analysis of Lens culinaris. J Hered. 77(6):473–474. doi:10.1093/oxfordjournals.jhered.a110287
- Mishra SK, Sharma B, Sharma SK. 2007. Genetics and cytogenetics of lentil. In: Yadav SS, McNeil DL, Stevenson PC, editors. Lentil: an ancient crop for modern times. Dordrecht: Springer; p. 187–208. doi:10.1007/978-1-4020-6313-8
- Naithani SP, Sarbhoy RK. 1973. Cytological studies in Lens esculenta Moench. Cytologia. 38(2):195–203. doi:10.1508/cytologia.38.195
- Nandanwar RS, Narkhede MM. 1991. Intraspecific variation in karyotype of lentil. J Maharastra Agri Univ. 16:24–27.
- Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 258(1):39–48. doi:10.1007/s00606-005-0389-2
- Sokal RR, Rohlf FJ. 1995. Biometry: the principles and practice of statistics in biological research. 3rd ed. New York (NY): W. H. Freeman & Co.
- Sharma AK, Muhkopadhyay S. 1963. Karyotype constancy in different strains of Lens esculenta Moench as worked out through recent techniques. Indian Agric. 7:103–111.
- Sinha SSN, Acharia SS. 1972. Karyotype analysis in some varieties of Lens culinaris. Cytologia. 37(4):673–683. doi:10.1508/cytologia.37.673
- van Oss H, Aron Y, Ladizinsky G. 1997. Chloroplast DNA variation and evolution of the genus Lens Mill. Theor Appl Genet. 94(3):452–457. doi:10.1007/s001220050436
- Weiss‐Schneeweiss H, Schneeweiss GM. 2013. Karyotype diversity and evolutionary trend in angiosperms. In: Leitch IJ, Greilhuber J, Dolezel J, Wendel JF, editors. Plant genome diversity, vol 2, physical structure, behavior and evolution of plant genomes. Vienna: Springer Verlag; p. 209–230.
- Zarco CR. 1986. A new method for estimating karyotype asymmetry. Taxon. 35(3):526–530. doi:10.2307/1221906