Abstract
The species Hovenia dulcis, originally from Asia, was introduced in several Brazilian regions due to its ornamental features, edible fruits and medicinal properties. Despite its importance, few cytogenetic studies have been carried out with this species. Literature data present chromosome counts for H. acerba and H. dulcis (2n = 24) and propose the basic chromosome number x = 12 for Hovenia, with no karyomorphological information. In this way classical and cytomolecular studies were carried out in H. dulcis. This study presents the first meiotic analysis, the karyotype and the first cytomolecular analysis for a Rhamnaceae species. The meiotic analysis showed a regular process and a high rate of viable pollen grains (91.3%). The chromosome numbers were n = 12 and 2n = 24, reinforcing the basic number proposed for the genus. The chromosomes are small (1.5–2.8 μm) and metacentric and the karyotype is symmetric. Two CMA+ bands at secondary constriction of the longest chromosome were identified. One pair of 45S rDNA sites in distal position and one pericentromeric pair of 5S rDNA sites were observed. The stable chromosome number and the single pair of rDNA sites indicate the absence of neopolyploidy in karyotype evolution of H. dulcis.
Introduction
The genus Hovenia Thunb. (tribe Paliureae – Rhamnaceae) has three species and two varieties naturally distributed in Korea, China and Japan. The exotic Hovenia dulcis, known as uva-do-japão in Brazil, is a deciduous tree that was introduced in several Brazilian regions due to its valuable wood, ornamental features and edible fruits (Yang et al. Citation2013). Moreover recent studies have pointed out some medicinal properties for its extracts, such as the significant reduction of liver injuries caused by alcohol in mice (Guo et al. Citation2015). Despite the large use of H. dulcis few studies have been carried out in this species, mainly focused on pharmacological usages and wood anatomy aspects.
Cytogenetic studies support biosystematic analysis presenting chromosomal characteristics and variation among the different taxa. This variation includes differences in the haploid and diploid chromosome numbers, chromosome morphology, inner chromosome structure and DNA sequence (Ilnicki Citation2014). The meiotic analysis may elucidate the genomic behavior during sporogenesis and reveal chromosome mutations. This analysis allows the valuation of reproductive viability of a population, and gives support for managing programs of plants of economic interest, as presented by Souza et al. (Citation2015) for Brachiaria hybrids.
The classical and cytomolecular techniques have been used to characterize the mitotic karyotypes. Classical assays staining permit chromosome accountings and karyological measurements. To refine the karyotype characterization, CMA/DAPI fluorochrome banding and in situ localization of specific sequences increase the cytogenetic data and improve chromosome idiograms. The fluorochrome chromomycin A3 (CMA) shows GC-rich sequences of DNA, and 4′,6-diamidine-2-phenylindole (DAPI) shows AT-rich DNA sites (Schweizer Citation1976).
The use of fluorescence in situ hybridization (FISH), with markers that identify specific chromosomes, provides a powerful approach to karyotype characterization and to genomic changes analysis within groups (Chester et al. Citation2010). The in tandem repeated sequences 45S rDNA, which represents the nucleolar organizing region (NOR), and 5S rDNA are both highly conserved in plants (Schubert Citation2007). These sequences have been used as probes in cytogenetic assays with remarkable success in cytogenetic characterization and in phylogenetic analysis, as presented by Yagi et al. (Citation2015) for Cucumis species.
The cytological literature presents the diploid chromosome number 2n = 24 for H. acerba and H. dulcis (Gadella et al. Citation1969; Oginuma et al. Citation1994, respectively), with no data for H. trichocarpa, the third species of Hovenia. Based on these counts those authors inferred the basic chromosome number x = 12 for the genus Hovenia. There are no references to meiotic process and karyomorphological data in the literature.
Cytogenetic studies were carried out in a H. dulcis population collected at municipality of Santo André, located at São Paulo metropolitan region, in southeastern Brazil. The objective of the present study was to analyze the male meiotic process (microsporogenesis), estimate pollen viability, report haploid and diploid chromosome number and present the first karyomorphological information for a Hovenia species, using CMA/DAPI banding and FISH with rDNA probes.
Materials and methods
Plant material and chromosome preparations
The plant material was collected at urban areas of Santo André, in São Paulo metropolitan region, southeastern Brazil. Vegetative and reproductive samples of Hovenia dulcis with collector number RAL089 were herborized and placed in the Universidade Federal do ABC herbarium. For the meiotic analyses flower buds were collected and fixed in Carnoy’s solution (alcohol 3:1 acetic acid, v/v). Excised anthers at different stages of development were squashed in 1.2% acetocarmine. For the estimation of pollen viability Alexander’s staining protocol (Citation1980) was followed.
For mitotic analyses seeds were collected and germinated in Petri dishes with distilled water at 25°C. The root tips were excised and pre-treated with the anti-mitotic agent PDB (p-dichlorobenzene) in saturated solution at 16°C for 3 h. The material was fixed in Carnoy’s solution for 24 h and stored at –20°C. For slide preparations root tips were digested in a solution of citrate buffer with 2% of cellulase and 20% of liquid pectinase for 1 h at 37°C and squashed in 45% acetic acid. For the idiogram construction chromosomes of 10 well-spread cells were stained with 2% Giemsa, measured with Micromeasure 3.3 software (Reeves & Tear, Citation2000) and classified following Levan et al.’s (Citation1964) nomenclature. The total chromatin length (TCL) of the diploid complement and the karyotype asymmetry rate TF% (Huziwara Citation1962) were estimated.
Fluorochrome banding and in situ hybridization
The protocols of Schweizer (Citation1976) were used for DAPI and CMA banding. The protocol of Pendas et al. (Citation1993) was used for FISH. The probes used were pTa71 which contains one unit of 45S rDNA (9 kb) derived from Triticum aestivum (Gerlach & Bedbrook Citation1979), and pScT7, with an insert of 5S rDNA with 300–500 bp isolated from Secale cereale (Lawrence & Appels Citation1986). The probe pTa71 was labeled with biotin-16-dUTP, and pScT7 with digoxigenin-11-dUTP (Boerhinger Mannheim, Germany) by nick translation, following the manufacturer’s instructions. The hybridized sites were detected with Avidin-Rhodamine and IgG-FITC respectively. The slides were counterstained with DAPI and mounted with Vectashield anti-fading (Vector Laboratories, USA). Slides were examined with a Nikon 80i epifluorescent microscope, Japan. Chromosome images were captured by a Nikon DS-Ri1 camera and processed in Nis-Elements software (Nikon Instruments, Japan), only using contrast and lightning adjustments, functions that affect the whole image equally.
Results
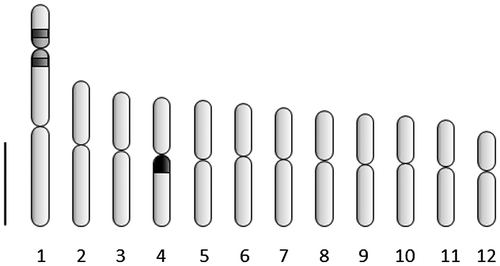
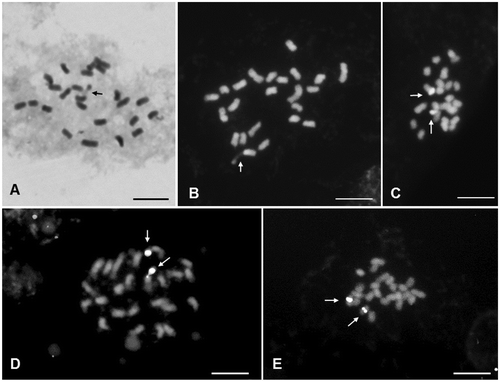
The haploid chromosome number n = 12 and the diploid count 2n = 24 were observed (Figure (A–C)). In meiotic analysis 12 bivalents were observed at diakinesis and metaphase I of meiosis, with no irregular structure, leading to 91.3% of pollen viability (Figure (D)). The mitotic chromosomes in metaphase are small (1.5–2.8 μm) and all metacentric (Table ). One pair of satellites was observed at chromosome 1 (Figure (A), 2(B)). The total chromatin length (TCL) is 47.78 ± 5.59 μm. The asymmetrical rate (TF%) was 48.8 indicating a symmetric karyotype. No DAPI+bands were observed (Figure (B)). One pair of CMA+bands was registered at distal positions of the longest chromosome (Figure (C)). The FISH with 45S rDNA probe indicated a secondary constriction in the short arm of pair 1 (Figure (D)), and the same region was stained with CMA banding. The two 5S rDNA sites are localized at the pericentromeric position of the long arm of pair four, not in synteny with the 45S rDNA sites (Figure €). The chromosome idiogram is presented in Figure . The karyotype formula is 2n = 24 = 12 m.
Figure 1. Pollen mother cells of H. dulcis in meiotic division. (A) diakinesis with 12 bivalents; (B) diakinesis with arrow showing the chromosome associated with the nucleolus; (C) metaphase I with n = 12; (D) pollens stained with Alexander (Citation1980) staining with arrows showing unviable ones. Bar = 5 μm.
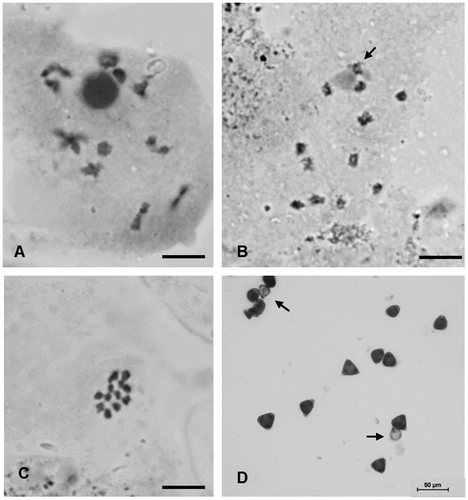
Table 1. Karyotype data of Hovenia dulcis, with average lengths of each pair of chromosomes (CL), their long arms (LA) and short arms (SA), centromeric index (CI) and chromosome classification (CC).
Figure 2. Cells of H. dulcis in metaphasic mitotic division. (A) Giemsa stained cell with 2n = 24 with arrow showing satellite; (B) DAPI stained cell with arrow showing satellite; (C) CMA stained cell with arrows showing positive bands; (D) cell counterstained with DAPI hybridized with pTa71 probe with arrows showing 45 rDNA sites; (E) cell hybridized with pScT7 probe with arrows showing 5S rDNA sites. Bar = 5 μm.
Discussion
The haploid chromosome number n = 12, unpublished for the species, and the diploid count 2n = 24 reinforces the x = 12 basic number proposed for the genus Hovenia. The tribe Paliureae of Rhamnaceae comprises three genera: Hovenia, Paliurus and Ziziphus (Richardson et al. Citation2000a). Chromosome number counts for species of these genera indicate the basic number x = 12 for the tribe. Only some Ziziphus species present chromosome counts divergent from this basic number, such as 2n = 22 for Z. joazeiro (Guerra 1986) and n = 11 for Z. mistol (Bernardello et al. Citation1990), indicating an x = 11 basic number. These divergent chromosome numbers may denote a descending disploidy in the karyotype evolution of the genus Ziziphus. In the descending disploidy the chromosome number variation is promoted by breakage of two chromosomes, followed by fusion of their broken ends. On the other hand the ascending disploidy is characterized by chromosome fission followed by the addition of telomere repeats to the broken termini (Guerra Citation2016). Disploidy seems to be an important process of karyotype divergence within Rhamnaceae. The tribe Gouanieae, which is the closest tribe phylogenetically to Paliureae (Richarson et al. Citation2000b), also presents counts different from x = 12, e.g. n = 23 registered for Gouania gagnei (Carr Citation1978) and n = 11 for Helinus lanceolatus (Khatoon and Ali Citation1993).
Polyploid accounts in Paliureae are restricted to Ziziphus species, e.g. the n = 24 observed by Sandhu and Mann (Citation1988) for Z. jujuba and the 2n = 36 presented by Khatoon and Ali (Citation1993) for Z. nummularia and Z. spina-christi. Besides this chromosome number variation, most of the data for Ziziphus are x = 12, indicating a numerical stability for the tribe Paliureae.
The high pollen viability with no chromosomal abnormalities observed here indicates a stable meiotic process in H. dulcis. This is the first data of pollen viability for a Hovenia species. Gotelli et al. (Citation2015) presented details of H. dulcis pollen and microsporangium development with no reference to pollen viability or meiotic index.
The colocalization of CMA+bands and 45S rDNA sites observed here is extensively reported in plant cytogenetics and seems to be a pattern due to the NOR heterochromatin associated characteristics (Guerra Citation2000). The 45S and 5S rDNA sites are not in synteny in H. dulcis. The syntenic localization of rDNA 45S and 5S sites is not very common and is probably a result of chromosomal rearrangements during karyotype evolution, as reported by Gan et al. (Citation2013) for the genus Gossypium.
The FISH assays with rDNA probes showed one pair of sites each in H. dulcis karyotype. The number of rDNA sites may not be strictly related to the ploidy level of a taxon, due to deletions, duplications and translocations of chromosome segments during evolution. Rosato et al. (Citation2015) presented a phylogenetic study on Vellinae group (Brassicaceae) based on chromosome and rDNA site numbers. Their results suggested independent changes in the rDNA loci number during speciation. However, as the basal number of rDNA sites is always one pair, multiple accountings may be accepted as a clue of genome polyploidization. In this way the presence of only one pair of 45S and 5S rDNA sites observed here may indicate the absence of a neopolyploidy in Hovenia dulcis karyotype evolution.
Based on the karyomorphological data presented here the first chromosome idiogram for the genus Hovenia was constructed. Karyotype data for Rhamnaceae genera were reported only for Karwinskia species (tribe Rhamneae) presenting small and mainly metacentric chromosomes. Tapia-Pastrana et al. (Citation2002) found 2n = 24 for K. humboldtiana, with karyotype formula 8 m + 4sm, and also observed one pair of satellites in mitotic metaphase. For K. mollis, K. tehuacana and K. umbellata Tapia-Pastrana et al. (Citation2004) presented also 2n = 24, with chromosome lengths from 0.68 to 1.28 μm and the predominance of metacentric ones. The chromosome satellites were all localized at the shorter chromosome pair in these three Karwinskia species, which is divergent from the secondary constriction localization observed here for H. dulcis. These data reinforce the role of chromosomal rearrangement, with few chromosome number variations, in Rhamnaceae karyotype evolution. The lack of karyomorphological data for Hovenia species shows the importance of this approach for evolutionary and systematic analysis, especially in groups with little or no chromosome number variation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported through a research grant from the Fundação de Amparo à Pesquisa do Estado de São Paulo - FAPESP [Proc. 04/14769-5].
References
- Alexander MP. 1980. A versatile stain for pollen fungi, yeast and bacteria. Stain Technol. 55:13–18. 10.3109/10520298009067890
- Bernardello LM, Stiefkens LB, Piovano MA. 1990. Números cromosómicos en dicotiledóneas Argentinas. Bol Soc Argent Bot. 26:149–157.
- Carr GD. 1978. Chromosome numbers of Hawaiian flowering plants and the significance of cytology in selected taxa. American Journal of Botany. 65:236–242.
- Chester M, Leitch AR, Soltis PS, Soltis DE. 2010. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridization). Genes. 1:166–192. 10.3390/genes1020166
- Gadella TW, Kliphuis E, Lindeman JC, Mennega EA. 1969. Chromosome numbers and seedling morphology of some Angiospermae collected in Brazil. Acta Bot Neerl. 18:74–83. 10.1111/plb.1969.18.issue-1
- Gan Y, Liu F, Chen D, Wu Q, Qin Q, Wang C, Li Shaohui. 2013. Chromosomal locations of 5S and 45S rDNA in Gossypium genus and its phylogenetic implications revealed by FISH. PLoS ONE. 8(6):e68207. 10.1371/journal.pone.0068207
- Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 7:1869–1885. 10.1093/nar/7.7.1869
- Gotelli MM, Galati BG, Zarlavsky G, Medan D. 2015. Pollen and microsporangium development in Hovenia dulcis (Rhamnaceae): a different type of tapetal cell ultrastructure. Protoplasma. 253:1125–1133.
- Guerra M. 2000. Patterns of heterochromatin distribution in plant chromosomes. Genet Mol Biol. 23:1029–1041. 10.1590/S1415-47572000000400049
- Guerra M. 2016. Agmatoploidy and symploidy: a critical review. Genet Mol Biol. 39(4):492–496. 10.1590/1678-4685-gmb-2016-0103
- Guo J, Meng Y, Zhao Y, Hu Y, Ren D, Yang X. 2015. Myricetin derived from Hovenia dulcis Thunb. ameliorates vascular endothelial dysfunction and liver injury in high choline-fed mice. Food Funct. 6:1620–1634. 10.1039/C4FO01073F
- Huziwara Y. 1962. Karyotype analysis in some genera of Compositae. VIII. further studies on the chromosomes of Aster. Am J Bot. 49:116–119. 10.2307/2439026
- Ilnicki T. 2014. Plant biosystematics with the help of cytology and cytogenetics. Caryologia. 67:199–208. 10.1080/00087114.2014.931642
- Khatoon S, Ali S. 1993. Chromosome atlas of the angiosperms of Pakistan. University of Karachi: Department of Botany, BCC&T Press; p. 232.
- Lawrence GJ, Appels R. 1986. Mapping the nucleolus organizer region, seed protein loci and isozyme loci on chromosome 1R in rye. Theor Appl Genet. 71(5):742–749.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Oginuma K, Gu Z, Yue Z, Kondo K. 1994. Chromosomes of some woody plants native to Yunnan, China. Kromosomo. 73:2491–2497.
- Pendas AM, Morán P, Garcia-Vázquez E. 1993. Ribosomal genes are interspersed throughout a heterochromatic arm in Atlantic salmon. Cytogenet Cell Genet. 63:128–130.
- Reeves A, Tear J. 2000. MicroMeasure for Windows, version 3.3. Free program distributed by the authors over the Internet from https://www.colostate.edu/Depts/Biology/MicroMeasure.
- Richardson JE, Fay MF, Cronk QCB, Chase MW. 2000a. A revision of the tribal classification of Rhamnaceae. Kew Bulletin. 55:311–340.
- Richardson JE, Fay MF, Cronk QC, Bowman D, Chase MW. 2000b. A phylogenetic analysis of Rhamnaceae using rbcL and trnL-F plastid DNA sequences. American Journal of Botany. 87(9):1309–1324.
- Rosato M, Moreno-Saiz JC, Galián JA, Rosselló JA. 2015. Evolutionary site-number changes of ribosomal DNA loci during speciation: complex scenarios of ancestral and more recent polyploid events. AoB Plants. 7: plv135.10.1093/aobpla/plv135
- Sandhu PS, Mann SK. 1988. SOCGI plant chromosome number reports VII. J Cytol Genet. 23:219–228.
- Schubert I. 2007. Chromosome evolution. Curr Opin Plant Biol. 10:109–115. 10.1016/j.pbi.2007.01.001
- Schweizer D. 1976. Reverse fluorescent chromosome banding with chromomycin and DAPI. Chromosoma. 58:307–324. 10.1007/BF00292840
- Souza VF, Pagliarini MS, Valle CB, Bione NCP, Menon MU, Mendes-Bonato AB. 2015. Meiotic behavior of Brachiaria decumbens hybrids. Genet Mol Res. 14:12855–12865. 10.4238/2015.October.21.5
- Tapia-Pastrana F, Mercado-Ruaro P, López S, Gómez A. 2002. Estudio citogenético en Karwinskia humboldtiana (Rhamnaceae) del Valle de Actopan, Hidalgo, México. Ann. Inst. Biol. Ser Bot. 73:17–25.
- Tapia-Pastrana F, Mercado-Ruaro P, Acevedo S, Nava R. 2004. Estudio cromosómico en tres especies de Karwinskia (Rhamnaceae) endémicas de México. Ann. Inst. Biol. Ser Bot. 75:1–10.
- Yagi K, Pawelkowicz M, Osipowski P, Siedlecka E, Przybecki Z, Tagashira N, Hoshi Yoshikazu. 2015. Molecular cytogenetic analysis of Cucumis wild species distributed in southern Africa: physical mapping of 5S and 45S rDNA with DAPI. Cytogenet Genome Res. 146:80–87. 10.1159/000433572
- Yang J, Wu S, Li C. 2013. High efficiency secondary somatic embryogenesis in Hovenia dulcis Thunb. through solid and liquid cultures. Sci World J. 2013:6. Article ID 718754.