Abstract
A karyological study of 53 populations of four Aegilops species (A. geniculata, A. triuncialis, A. ventricosa and A. neglecta) sampled in different eco-geographical sites in the north of Algeria was carried out using the Feulgen staining method. Chromosome counting showed that three species, A. geniculata, A. triuncialis and A. ventricosa, were tetraploid (2n = 4x = 28), whereas A. neglecta is hexaploid (2n = 6x = 42). Karyo-morphometric analysis of somatic metaphase chromosomes revealed four cytotypes in A. geniculata and two cytotypes in A. triuncialis, A. ventricosa and A. neglecta. The cytotypes found within the studied populations are differentiated by the lengths of the chromosomes, the symmetry of karyotypes and the presence or absence of the satellites. This difference indicates the relationships among Aegilops species by absence of telocentric chromosomes in any karyotype formula. However, chromosomes pairings in diakinesis, metaphase I and anaphase I revealed a less regular meiosis of tetraploid and hexaploid samples, with univalent, chromosomes lagging and chromatin bridges anomalies. Finally, results show the high genetic diversity of Aegilops populations from Algeria, suggesting that environmental factors and geographical location might have an effect on the genetic structure and evolution of chromosomes, which is of interest for cytogenetic identification and characterization of populations, to assess and enhance plant genetic resources.
Introduction
The genus Aegilops L, belonging to family Poaceae, tribe Triticeae Dumort., subtribe Triticineae Fr., includes 22 annual self-fertile species (Van Slageren Citation1994), representing the main genetic reserve for the improvement of wheat cultivars (Kilian et al. Citation2011). These species have a wide distribution in Asia, the Mediterranean basin and in North America (Sakamoto and Kobayashi Citation1980), showing adaptations to a large range of environmental constraints reflecting a large morphological variability of populations. Several biosystematic studies have clarified the taxonomic status and assessed the genetic resources of these species (Zhukovsky Citation1928; Eig Citation1929; Sears Citation1941; Kihara Citation1946; Hammer Citation1980; Kimber and Feldman Citation1987; Van Slageren Citation1994; Kilian et al. Citation2011; Giraldo et al. Citation2016).
Because of their strong phylogenetic relationships with species of Triticum, many taxonomic problems and nomenclature are posed. According to Van Slageren (Citation1994), the genus is divided into five sections: Aegilops, Comopyrum, Cylindricum, Sitopsis and Vertebrata and it consists of 11 diploid species (2n = 2x = 14), 10 tetraploid species (2n = 4x = 28), and two hexaploid species (2n = 6x = 42). However, recent studies based on molecular characters such as two single-copy nuclear genes (DMC1 and EF-G), showed that Aegilops speltoides, Triticum urartu and Aegilops tauschii provided respectively the B, A and D genome of cultivated wheat, suggesting that Aegilops and Triticum are monophyletic (Petersen et al. Citation2006). Also, the evolutionary history in a reticulated manner, of two genera Aegilops and Triticum within Triticeae is obtained for the first time, using the supermatrix and Bayesian concordance factor (BCF) approaches for construction of multigenic network including one chloroplastic and 26 nuclear genes (Escobar et al. Citation2011).
In Algeria, the genus Aegilops is represented by A. ventricosa Tausch, A. peregrina Rack and A. triuncialis L. subsp. ovata Eig (= A. geniculata Roth), subsp. triaristata (Willd.) Rouy (= A. neglecta Req. ex Bertol) and subsp. eu-triuncialis Eig (Quezel and Santa Citation1962). Populations of Aegilops geniculata show a wide range of adaptation to different bioclimatic conditions and this species is represented in very diverse habitats, from coastal full, the Tell Atlas, the high steppe plains to the Saharan Atlas, while the other species are characterized by scattered populations and have a limited distribution (Bandou et al. Citation2009).
Cytogenetic studies of Aegilops clarified the systematic and phylogenetic position of some taxa (Senjaninova – Korchagina Citation1932; Chennaveeraiah Citation1960; Kimber and Feldman Citation1987; Van Slageren Citation1994), highlighting that the diploid species can be grouped in seven genome types: C (A. markgrafii), M (A. comosa), N (A. uniaristata), D (A. tauschii), U (A. umbellulata), T (A. mutica) and S (A. speltoides, A. bicornis, A. longissima, A. sharonensis and A. searsii), whereas the polyploid species resulted from different combinations of these genomes. Recently, Badaeva et al. (Citation2002a, 2002b, Citation2004, Citation2011) established phylogenic relationships between the polyploid species and their diploid parents by C-banding and cytomolecular techniques. However, other molecular approaches have been developed to obtain better knowledge of the phenomena of speciation and evolution in these species, e.g. the study of reserve proteins (Fernández-Calvín and Orellana Citation1990; Rodriguez-Quijano et al. Citation2000; Sun et al. Citation2006) and DNA polymorphism (Zhang et al. Citation1996; Zaharieva et al. Citation2001; Sasanuma et al. Citation2004; Salina et al. Citation2006; Haider and Nabulsi Citation2008; Mahjoub et al. Citation2010; Giraldo et al. Citation2016).
The main objective of this study was to evaluate the genetic diversity of Aegilops populations sampled in a wide range of ecological conditions in north Algeria. In order to understand the role of ecological factors in the differentiation and evolution of chromosomes, we followed a cytogenetic approach based on karyotype characteristics and chromosome behavior. Furthermore, our results could be useful to improve the management and conservation of genetic resources of these species.
Materials and methods
Collection and sampling
Thirty-five populations of Aegilops geniculata, eight Aegilops neglecta, six Aegilops ventricosa and four Aegilops triuncialis were sampled from May 2012 to July 2013 according to an east–west rainfall gradient and north–south aridity gradient in north Algeria (Figure ). Each sampling site is characterized by the main ecological factors of the Mediterranean climate, i.e. the average annual rainfall (P), the average temperature of the hottest month in summer (M), the average temperature of the coldest month in winter (m), and altitude (Alt) used in the bioclimatic coefficient (Q2) (Table ). In each sampling site, one spike per plant of 10 complete individuals randomly collected (each separated by at least 1 m), was taken for chromosome counting and karyological analysis.
Figure 1. Location of sampled populations of four species of Aegilops in Algeria (g = A. geniculata, n = A. neglecta, v = A. ventricosa and t = A. triuncialis). Bioclimatic limits according to Stewart (Citation1974).
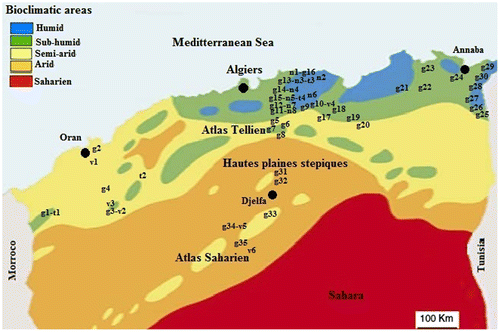
Table 1. Climatic and floristic characteristics of sampling sites of natural populations of Aegilops in Algeria.
Chromosome counting
Chromosome counts were performed on metaphase somatic cells in root meristems, according to the Feulgen method (Jahier et al. Citation1992). After germination of caryopses at room temperature (20°C) for 48 h, the roots (1–1.5 cm) were cut and processed by an α-bromonaphthalene saturated solution at room temperature for 2 h. The fixing was done in a mixture of alcohol–acetic acid (3:1) at room temperature for 24 h and then preserved in 70° alcohol at 4°C. The cut roots were subsequently rinsed in distilled water and hydrolyzed by 1 N HCl at 60°C for 10 min before being stained in Schiff reagent for 1–2 h in the dark. After rinsing with water to remove excess dye, the root ends stained bright red were cut and mounted between slide and cover in a drop of acetic carmine or in acetic acid 45% to increase the contrast of the chromosomes and the cytoplasm. Then a squash was made which consists of preparations crushed with a match. Observations were made by light microscopy at 40 × magnification. Good quality metaphase plates which have well individualized chromosomes were photographed in a Zeiss microscope Axiostar (Zeiss, Gottingen, Germany) with a digital camera.
Karyological analysis
The karyological analysis is based on average measures of the length of short arm (Bc), long arm (Bl) and total length (Lt) of spread chromosomes in a minimum of three best metaphase plates, using karyological software (Ideokar 1.2 software package version Citation2015). The average of total lengths of all chromosomes (Χ), the ratio of long arm and short arm (R) and centromeric index (CI) were calculated. The karyotypes are established based on the characteristics of the chromosomes according to the number, form, size and secondary construction or satellite. They are described following the nomenclature of Levan et al. (Citation1964). Idiograms were drawn and arranged from left to right by size in decreasing order of average length of chromosomes and the centromeric index.
Karyotype asymmetry was estimated by using the coefficient of variation of chromosome length (CVCL) (Paszko Citation2006), which gives a measure of interchromosomal asymmetry, and the mean centromeric asymmetry (MCA) (Peruzzi and Eroglu Citation2013), which gives a measure of intrachromosomal asymmetry. For correlations among karyological characters and environmental factors, the Pearson coefficient is performed. Meanwhile, to infer karyological relationships among the four studied species, the standardized approach proposed by Peruzzi and Altınordu (Citation2014) was used; this method focuses on principal coordinate analysis (PCoA) as a general ordination method, integrating six different, not redundant, karyological parameters: basic chromosome number (x), chromosome number (2n), total haploid chromosome length (THL), mean centromeric asymmetry (MCA), coefficient of variation of chromosome length (CVCL) and coefficient of variation of centromeric index (CVCI). To perform these analyses, Software Past 3.15 (Hammer et al. Citation2001; Hammer Citation2017) was used.
Pollen meiosis
The chromosome pairing was analyzed at the pollen mother cells (PMC) during diakinesis (prophase I), metaphase I and anaphase I in populations of Aegilops geniculata and neglecta by Feulgen technique (Jahier et al. Citation1992). The spikes collected before anthesis were fixed in Carnoy solution (ethanol-chloroform-acid acetic 6:3:1 v/v/v), for 48 h at room temperature. Conservation was carried out in 70% ethanol at 4°C. After acid hydrolysis at 60°C the young anthers were dissected and mounted between slide and cover in a carmine acetic drop, then a squash technique was done. Observations were performed using a microscope with a digital camera.
Afterwards, the number of rod bivalent, ring bivalent and anomalies such as univalents, chromosomes lagging and chromatin bridges, were counted to determine the meiotic configurations at metaphase I (MI. configuration).
Results
Chromosome counting
Chromosome counts performed on 258 metaphase plates in Aegilops geniculata, 112 in A. neglecta, 45 in A. triuncialis and 42 in A. ventricosa (Table ) showed the existence of two main chromosome numbers: 2n = 4x = 28 for the populations of A. geniculata, A. triuncialis and A. ventricosa and 2n = 6x = 42 for A. neglecta. Other numbers (2n = 27, 29, 39, 40, 41) were found in the same cells at low frequencies, which may be due to observation errors or aneuploidy phenomena.
Table 2. Number of individuals, mitotic cells, absolute frequencies (FA) and relative frequencies (FR) of the chromosome numbers found in the populations of Aegilops from Algeria.
Cytotype and idiogram characters
The different karyo-morphometric parameters (Table ) performed on all spread chromosomes from the best metaphase plates with 2n = 4x = 28 and 2n = 6x = 42 (Figures ) allowed identification of four tetraploid cytotypes in Aegilops geniculata, two tetraploid cytotypes (1 and 2) for A. triuncialis and two for A. ventricosa, and two hexaploid cytotypes for A. neglecta. The idiograms (Figure ) are established from the mean of measures taken on the chromosomes of three somatic metaphase of the cytotypes found in four species.
Table 3. Chromosome number (2n), karyotypes formulas, intrachromosomal (A1, CVci), and interchromosomal (A2, CVcl) asymmetry indices and the means values of the total length of chromosomes (Χ ± ES) of four species of Aegilops from Algeria.
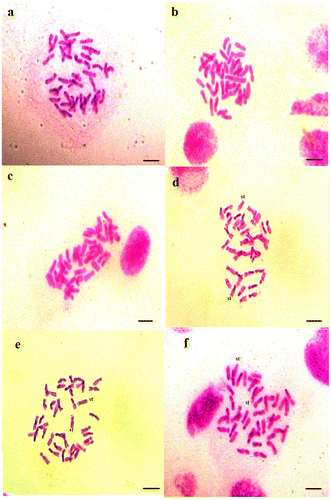
Figure 2. Micrographies of tetraploid metaphase plates (2n = 4x = 28) of Aegilops geniculata. (a–c) Cytotype 1; (d–f) Cytotype 2; (g–i) Cytotype 3; (j–l) Cytotype 4. st: satellite. Scale bar = 15 μm.
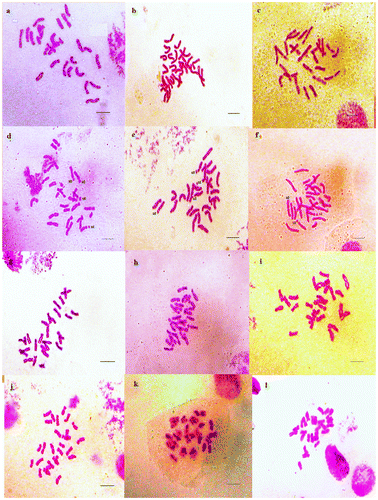
Figure 3. Micrographies of tetraploid metaphase plates (2n = 4x = 28) of Aegilops triuncialis. (a–c) Cytotype 1; (d–f) Cytotype 2. st: satellite. Scale bar = 15 μm.
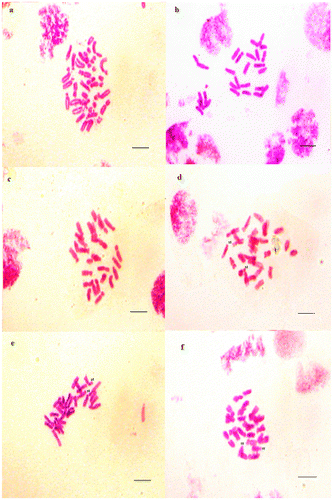
Figure 4. Micrographies of tetraploid metaphase plates (2n = 4x = 28) of Aegilops ventricosa. (a–c) Cytotype 1; (d–f) Cytotype 2. Scale bar = 15 μm.
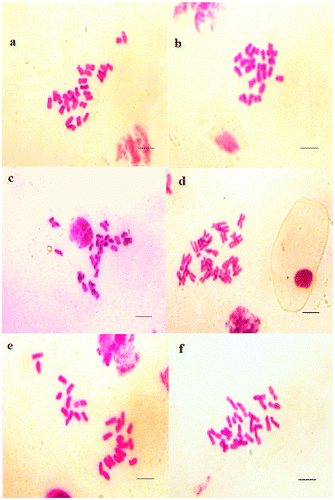
Figure 5. Micrographies of hexaploid metaphase plates (2n = 6x = 42) of Aegilops neglecta. (a–c) Cytotype 1; (d–f) Cytotype 2. st: satellite. Scale bar = 15 μm.
Aegilops geniculata
Cytotype 1 was found in populations sampled in coastal regions and the plain at an average altitude under a subhumid and humid bioclimate. It contains large chromosomes, with lengths of 12.93–31.48 μm and mean value of total length of 24.3 μm. The karyotype formula consists of seven metacentric chromosome pairs, four submetacentric chromosome pairs and three subtelocentric pairs. Cytotype 2 was found in populations of plains and mountains at an average altitude under a sub-humid and semi-arid bioclimate; it contains chromosomes of 6.68–25.29 μm, with mean value of total length of 18.61 μm and the karyotype is composed of three metacentric chromosome pairs, 10 submetacentric chromosome pairs of which two have satellites, and one subtelocentric pair. Cytotype 3 was found in populations of the high steppe plains under an arid bioclimate. The chromosomes have intermediate length of 16.25–25.38 μm with a mean value of total length of 20.14 μm and its karyotype formula is formed by six metacentric chromosome pairs, four submetacentric chromosome pairs and four subtelocentric pairs. Cytotype 4 was found in populations sampled in mountains at high altitudes of the Tellian and Saharian Atlas under a humid and arid bioclimate. It contains small chromosomes of 5.65–18.93 μm with mean values of total length of 15.73 μm, and the karyotype is characterized by five metacentric chromosome pairs, six submetacentric chromosome pairs and three subtelocentric pairs.
Aegilops triuncialis
Cytotype 1 was found in populations sampled in plains at low and average altitudes under a humid bioclimate. It consists of chromosomes of 8.36–20.42 μm and an average total length of 14.59 μm. These populations have karyotype formula of seven metacentric chromosome pairs, four submetacentric chromosome pairs and three subtelocentric pairs.
Cytotype 2 was found in populations of high steppe plains under a semi-arid bioclimate. It is composed by smaller chromosomes with lengths ranging from 6.46 to 14.47 μm, and an average total length of 11.25 μm. The karyotype formula has nine metacentric chromosome pairs, three submetacentric chromosome pairs of which one has a satellite, and two subtelocentric pairs.
Aegilops ventricosa
Cytotype 1 was found in populations of plains and mountains at an average altitude under a sub-humid and semi-arid bioclimate. It contains small chromosomes with lengths of 4.87–11.65 μm, with mean value of total length of 10.38 μm, and the karyotype is composed of 10 metacentric chromosome pairs, three submetacentric chromosome pairs and one subtelocentric pair.
Cytotype 2 was found in populations sampled in mountains at high altitudes of the Tellian and Saharian Atlas under a humid and arid bioclimate. It is characterized by large chromosomes with lengths of 12.61–21.68 μm and an average total length of 16.41 μm. This karyotype is characterized by six metacentric chromosome pairs, five submetacentric chromosome pairs and three subtelocentric pairs.
Aegilops neglecta
Cytotype 1 was found in all populations, and consisted of large chromosomes, with lengths of 9.75–20 μm and mean value of total length of 14.36 μm. The karyotype is characterized by seven metacentric chromosome pairs, nine submetacentric chromosome pairs and five subtelocentric pairs.
Cytotype 2 was found in populations of plains and mountains at an average altitude under a humid bioclimate. The chromosomes have intermediate size with lengths of 9.37–15.79 μm and a mean value of total length of 13.22 μm. These populations have 13 metacentric chromosome pairs, five submetacentric chromosome pairs of which one has a satellite, and two subtelocentric pairs.
Study of karyotypes asymmetry
Values of the mean centromeric asymmetry and interchromosomal asymmetry indices applied to the tetraploid and hexaploid cytotypes of four species of Aegilops are shown in Table .
According to the mean centromeric asymmetry (MCA), the tetraploid cytotypes (1, 3 and 4) of Aegilops geniculata, (1 and 2) of Aegilops triuncialis, (2) of Aegilops ventricosa and the hexaploid cytotypes (1 and 2) of Aegilops neglecta had the most asymmetrical karyotype, with values ranging from 21.96 to 33.50. Meanwhile, the tetraploid cytotype (2) of Aegilops geniculata and cytotype (1) of Aegilops ventricosa have low intrachromosomal karyotype asymmetry with which vary respectively from 15.24 to 18.03. Although the interchromosomal asymmetry indices (CVCL) does not show a difference between cytotypes of each four studied species, because the size of the chromosomes varies almost the same, Aegilops triuncialis has the higher interchromosomal karyotype asymmetry.
Infer karyological relationships among species
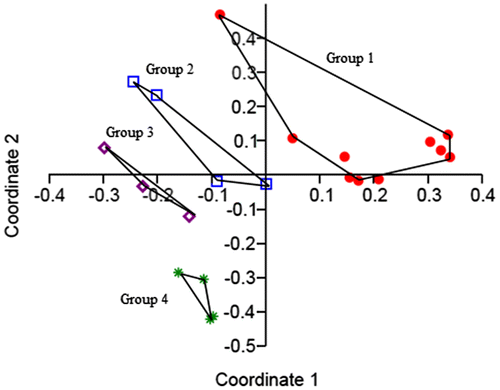
Multivariate analysis of karyological characters of populations sampled in the differentiated ecological sites (Figure ) resulted in a separation of karyotypes in four mainly groups. Group 1 is heterogeneous, composed by karyotypes of populations of A. geniculata; group 2 comprises karyotypes of populations of A. ventricosa; group 3 is made up of karyotypes of populations of A. triuncialis; and group 4 contains hexaploid karyotypes of populations of A. neglecta.
Figure 7. Principal coordinate analysis (PCoA) for Aegilops accessions based on six quantitative karyological parameters.
A Pearson coefficient among karyological characters and environmental factors (Table ) allowed us to determine the following correlations: THL is negatively correlated with altitude (Alt). CVCL is negatively correlated with the altitude (Alt) and independent of the highest temperature in summer (M) and bioclimatic coefficient (Q2). CVCI and MCA are strongly linked each other and positively correlated with annual rainfall (p), coldest temperature in winter (m) and bioclimatic coefficient (Q2). Lastly, 2n and x exhibit a low correlation with ecological parameters.
Table 4. Correlation table with Pearson coefficient, among karyological characters and environmental factors.
Meiotic analysis
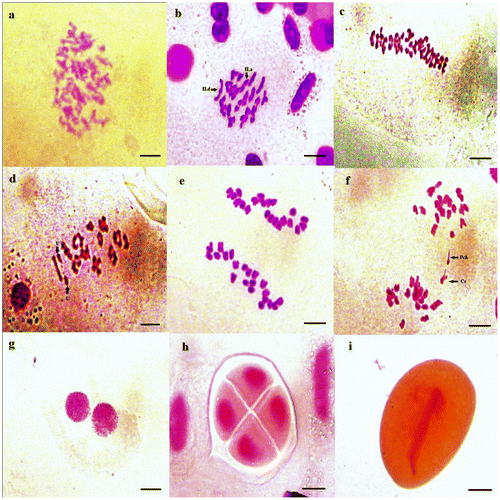
The chromosome pairing during diakinesis, metaphase I and anaphase I (Table ) showed that the meiosis in the tetraploid samples of Aegilops geniculata (Figure ) is regular presented by the high frequency of ring bivalents and MI configuration of 10II.a+4II.d. Yet, some individuals presented anomalies. Whereas, the meiosis in the majority of the hexaploid samples of Aegilops neglecta studied (Figure ), is irregular that were characterized by the abundance of rod bivalents with two MI configurations of 9II.a+12II.d and 10II.a+11II.d, and the presence of univalents, chromosome lagging and chromatic bridges anomalies at anaphase I.
Table 5. Number of mother pollen cells (MPC) and MI configurations frequencies of Aegilops geniculata and Aegilops neglecta.
Figure 8. Micrographs of pollen meiosis of Aegilops geniculata (2n = 4x = 28): (a) pachytene; (b) diakinesis; (c, d) metaphase I (U: univalent chromosomes); (e, f) anaphase I (Pch: chromatin bridges and Cr: chromosome lagging); (g) dyad; (h) tetrad; (i) pollen grain with pore aperture. Scale bar = 15 μm.
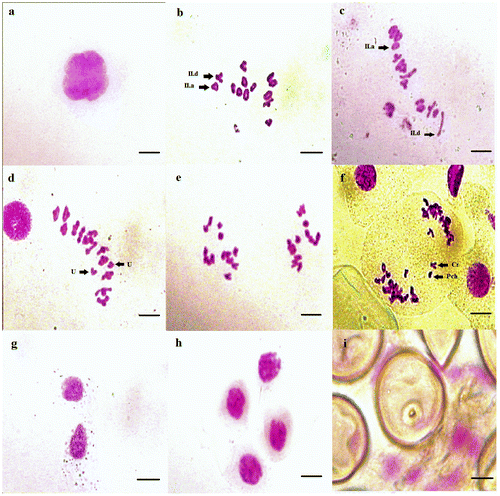
Figure 9. Micrographs of pollen meiosis of Aegilops neglecta (2n = 6x = 42): (a) pachytene; (b) diakinesis; (c, d) metaphase I (U: univalent chromosomes); (e, f) anaphase I (Pch: chromatin bridges and Cr: chromosome lagging); (g) dyad; (h) tetrad; (i) pollen grain with furrow aperture. Scale bar = 15 μm.
Discussion
The observation of the somatic mitoses in all investigated populations of the four species of Aegilops showed a tetraploid chromosome number of 2n = 4x = 28 in Aegilops geniculata, A. triuncialis and A. ventricosa, as highlighted by previous counts (Kihara Citation1954; Chennaveeraiah Citation1960; Kimber and Feldman Citation1987; Van Slageran Citation1994). Meanwhile, in populations of Aegilops neglecta only one hexaploid chromosomal number (2n = 6x = 42) was counted. Van Slageren (Citation1994) and Dvorak (Citation1998) reported two ploidy levels for this species corresponding to Aegilops neglecta Req. ex Bertol. subsp. neglecta as tetraploid (2n = 4x = 28) and Aegilops neglecta Req. ex Bertol. subsp. recta (Zhuk.) Chennav (2n = 6x =42) as hexaploid. However, we have not encountered any diploid individuals (2n = 2x = 14), confirming the allopatric distribution of these species which are characterized by a large distribution compared to their diploid parents, which are limited to their centers of origin in Asia (Kilian et al. Citation2011). According to several authors (Kihara Citation1954; Zohary Citation1966; Hammer Citation1980; Dubcovsky and Dvorak Citation1994; Badaeva E.D Citation2002a) the diploid species of the genus Aegilops are characterized by lower genetic variability than related polyploid species, which are characterized by high genetic variability and the ability to colonize new ecogeographical areas. We also confirm results that suggested an allopatric distribution of both tetraploid and hexaploid taxa of Aegilops neglecta, with more frequent hexaploids in the west and tetraploids in the east of the Mediterranean Sea (Belkadi et al. Citation2003; Ohta et al. Citation2015).
The analysis of metaphase chromosomes in tetraploid and hexaploid somatic cells by karyo-morphometric parameters allowed determination of the high karyological diversity revealed by a large difference of the length, the asymmetry of the chromosomes and presence or absence of satellite in cytotypes established among and between the four species studied. According to Stebbins (Citation1971), asymmetric karyotypes are either made of chromosomes similar in size with a shift in the position of the centromere from metacentric to telocentric, or composed of chromosomes different in sizes. In contrast, the karyological analysis of Aegilops crassa Boiss. from Iran (Hatami-Maleki et al. Citation2010), which has two tetraploid and hexaploid cytotypes, showed the presence of a single tetraploid chromosomal number of 2n = 4x = 28 and the karyological formulas of the different populations studied are similar, characterized by the abundance of metacentric chromosomes. However, populations are differentiated by some chromosomal structures such as the presence or absence of satellites. Badaeva et al. (Citation2002a, Citation2002b, Citation2004) using the C-banding technique and FISH, were able to establish the phylogenetic relationships among diploid species and allopolyploids and demonstrated the mechanisms of genomic differentiation within the same species.
The large intraspecific and interspecific variability of the chromosome length found in the different cytotypes of the four species of Aegilops sampled in north Algeria may be explained by the loss of genetic information by mutations such as fissions and deletions, which have beneficial effects on the evolutionary process and adaptation (Gorenflot and Raicu Citation1980). On the other hand, the increase in chromosome length would make it possible to adapt to eco-geographical conditions such as mountains of high altitudes and arid lands where some populations are sampled, thus allowing colonization of new ecological habitats (Levin Citation2002). In the case of Carthamus tinctorius L. the polymorphism of karyotypes and chromosomes is reflected by the variability in total length (short and long arm) of the chromosomes and there is no change in the karyotype formulas, which are symmetrical in all populations. This suggests that the evolution of karyotypes is related to changes in the amounts of genomic DNA that are proportional to the length of each chromosome arm (Anjali and Srivastava Citation2012). Therefore, the large karyotype polymorphism expressed by the variability of length and type of chromosomal found in Aegilops species studied can be explained by evolutionary processes and genome dynamics (Soltis and Soltis Citation1999). This structural diversity of chromosomes and karyotypes is often reflected in the phenotypic variability of several neopolyploid species (Ramsey and Schemske Citation2002), and this fact can favor adaptive evolutionary changes (Otto and Whitton Citation2000).
Other studies have attempted to correlate changes in chromosome numbers and patterns with eco-geographical conditions. Hence, in Turkey the species of Crocus (Uslu et al. Citation2012), Onobrychis Mill. and Hedysarum L. (Arslan et al. Citation2012) and Allium L. (Genç and Fırat Citation2016) sampled in the various ecological habitats show a great diversity in the structure of karyotypes, expressed by a large chromosomal number variability and polymorphism in chromosome structures such as size, and chromosomal types varying from metacentric to telocentric. In Algeria, caryological study of the genus Prospero L., Barnardia Poir. and Hyacinthoides Poir. of Asparagaceae (Hamouche et al. Citation2010), Allium of Amaryllidaceae (Khedim et al. Citation2015), Bellevalia and Muscari of Asparagaceae (Azizi et al. Citation2016), and Plantago of Plantaginaceae (Maamri et al. Citation2016) sampled under various ecogeographic conditions, showed intra- and inter-specific differences in ploidy level, chromosome structure, and karyotype asymmetry, which allowed the differentiation of the taxa studied and the clarification of their taxonomic status in the floras of Algeria (Maire Citation1955; Quezel and Santa Citation1962) compared to the new classification.
Finally, the multivariate approach (PCoA) was shown to be a good way to establish the karyological relationships among the four species of Aegilops collected in northern Algeria. The most important characters to recognize are the chromosome number (2n), total haploid chromosome length (THL), mean centromeric asymmetry (MCA) and coefficient of variation of chromosome length (CVCL). This result clarified the taxonomic status of A. triuncialis L. subsp. ovata Eig and Aegilops triuncialis L. subsp. triaristata (Willd.) Rouy in Algerian flora (Quezel and Santa Citation1962). Meanwhile, in recent classifications (Van Slageren Citation1994; APG III Citation2009), these species are separated respectively as: A. geniculata Roth and A. neglecta Req. ex Bertol. Also, this analysis was used by Peruzzi and Altınordu (Citation2014) to elucidate karyological affinity between several taxa as families (Liliaceae and Smilacaceae), tribes (Tulipeae, Lilieae, Medeoelae, Streptopeae, Tricyrtideae and Calochorteae), genera (Erythronium, Tulipa, Amana and Gagea), sections (Annui, Cyananthus, and Stenolobi) and related species of the genus Crocus. The Pearson correlation coefficient showed the influence of environmental factors on karyological characters. Significant correlations were found between other genetic characters of the Aegilops from Algeria, such as the glutenin subunits (Bandou et al. Citation2009) and gliadin subunits (Medouri et al. Citation2015), with environmental conditions.
Chromosomes analysis of Aegilops geniculata PMC revealed that chromosome pairing in metaphase I on the majority of samples is in ring bivalent, while in Aegilops neglecta is in rod bivalent, with some samples characterized by anomalies such as univalents, chromatin bridges and chromosomes lagging in anaphase I, due probably to inversion mutation (Sybenga Citation1975), reflecting an irregular meiosis in this species. We noted only bivalents and absence of multivalent in the pairing of homologous chromosomes, confirming allopolyploid (amphiploid) genesis of these species (Cunado Citation1992). Moreover, the chromosome behavior in meiosis of polyploid Aegilops species are scarce except the findings of Pignone et al. (Citation1994) on different varieties and sub species of Aegilops ventricosa that showed regular meiosis including bivalents and no abnormalities in this species. Indeed, our results allowed us to be more precise than Cunado (Citation1992) and Cuñado et al. (Citation1996), who reported that the polyploid Aegilops species have regular meiosis. On the other hand, several authors (Roy Citation1959; Cifuentes and Benavente Citation2009) established phylogenetic relationships between different species of Aegilops and Triticum genera by the analysis of the chromosome pairing in meiosis, mainly in metaphase I.
Conclusion
The current paper presents the somatic and meiotic chromosomal characteristics of four taxa of the genus Aegilops distributed in various eco-geographical areas in the north of Algeria. Our data show that Algerian Aegilops populations are polyploidy complex with impressive karyological diversity. This fact suggests that eco-geographical conditions might have an effect on changes in genetic structure of Aegilops populations. Furthermore, north Algeria has specific features, and its transitional position between two contrasting areas (the desert to the south and the Mediterranean Sea to the north) offers the opportunity to study evolutionary trends of species and natural populations. However, the present work sheds light on the role of ecological factors in the structure, function and evolution of chromosomes and interest for the use of cytogenetic techniques in the identification, to assess and enhance plant genetic resources.
Disclosure statement
The authors declare that they have no conflict of interest.
Acknowledgment
The present work has received financial assistance from the University of Sciences and Technology Houari Boumediene (USTHB, Algiers, Algeria). It was conducted in the framework of the program “The assessment of morphological and genetic diversity of spontaneous species in Algeria” of the Team Biosystematics, Genetics and Evolution (Project: Cnepru). The authors would thank the editor and reviewers for their comments and suggestions that considerably improve the manuscript.
References
- Anjali M, Srivastava A. 2012. Karyological studies in twelve accessions of Carthamus tinctoriusi. Caryologia. 65:1–6. doi:10.1080/00087114.2012.678072.
- APG III. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 161:105–121. doi:10.1111/j.1095-8339.2009.00996.
- Arslan E, Ertuğrul K, Tugay O, Dural H. 2012. Karyological studies of the genus Onobrychis Mill. and the related genera Hedysarum L. and Sartoria Boiss. & Heldr. (Fabaceae) from Turkey. Caryologia. 65 (1):11–17. doi:10.1080/00087114.2012.678079.
- Azizi N, Amirouche R, Amirouche N. 2016. Karyological investigations and new chromosome number reports in Bellevalia Lapeyrouse, 1808 and Muscari Miller, 1758 (Asparagaceae) from Algeria. Comp Cytogen. 10:171–187.
- Badaeva ED. 2002a. Evaluation of phylogenetic relationships between five polyploid Aegilops species of the U-genome cluster by means of chromosome analysis. Russian J Genetics. 38:799–811.
- Badaeva ED, Amosova AV, Muravenko OV, Samatadze TE, Chikida NN, Zelenin AV, Friebe B, Gill BS. 2002b. Genome differentiation in Aegilops. 3. evolution of the D-genome cluster. Plant Syst Evol. 231 (1-4):163–190. 10.1007/s006060200018
- Badaeva ED, Amosova AV, Samatadze TE, Zoshchuk SA, Shostak N, Chikida AV, Zelenin WJ, Raupp B, Friebe BS. 2004. Genome differentiation in Aegilops. 4. Evolution of U-genome cluster. Plant Syst Evol. 246:45–76.10.1007/s00606-003-0072-4
- Badaeva ED, Dedkova OS, Zoshchuk SA. 2011. Comparative analysis of the N-genome in diploid and polyploidy Aegilops species. Chromosome Res. 19(4):541–548.10.1007/s10577-011-9211-x
- Bandou H, Rodriguez-Quijano M, Carrillo JM, Branlard G, Zaharieva M, Monneveux P. 2009. Morphological and Genetic variation in Aegilops geniculata Roth, from Algeria. Plant Syst Evol. 277:85–97.10.1007/s00606-008-0106-z
- Belkadi B, Assali N, Benlhabib O. 2003. Variation of specific morphological traits and ploidy level of five Aegilops L. species in Morocco. Acta Bot Mal. 28:47–58.
- Chennaveeraiah MS. 1960. Karyomorphologic and cytotaxenoaie studies in Aegilops. Acta Horti Gotoburg. 23:85–178.
- Cifuentes M, Benavente E. 2009. Wheat-alien metaphase I pairing of individual wheat genomes and D genome chromosomes in interspeciWc hybrids between Triticum aestivum L. and Aegilops geniculata Roth. Theor Appl Genet. 119:805–813.10.1007/s00122-009-1090-6
- Cunado N. 1992. Analysis of Metaphse I chromosome association in species of genus Aegilops. Theor appl Genet. 85:283–292.
- Cunado N, Callejas S, Garela MJ, Fernfindez A, Santos JL. 1996. The pattern of zygotene and pachytene pairing in allotetraploid Aegilops species sharing the U genome. Theor appl Genet. 93:1152–1155.10.1007/BF00230139
- Dubcovsky J, Dvorak J. 1994. Genome origin of Triticum cylindricum, Triticum triunciale and Triticum ventricosum (Poaceae) inferred from variation in repeated nucleotide sequences: a methodological study. Am J Bot. 81:1327–1335.10.2307/2445408
- Dvorak J. 1998. Genome analysis in the Triticum-Aegilops alliance. In: Proceedings of the 9th international wheat genetics symposium. Saskatoon; p. 8–11.
- Eig A. 1929. Monographisch-kritische U¨ bersicht der Gattung Aegilops. Feddes Repert. 55:1–228.
- Escobar JS, Scornavacca C, Cenci A, Guilhaumon C, Santoni S, Douzery EJ, Ranwez V, Glémin S, David J. 2011. Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae). BMC Evol Biol. 11:568. doi:10.1186/1471-2148-11-181.
- Fernandez-Calvin B, Orellana J. 1990. High molecular weight glutenin subunit variation in the Sitopsis section of Aegilops. Implications for the origin of the B genome of wheat. Heredity. 65:455–463.10.1038/hdy.1990.117
- Genç I, Fırat M. 2016. Karyological studies of six taxa of the genus Allium L. (Amaryllidaceae) from Turkey. Caryologia. 69:187–190. doi: 10.1080/00087114.2016.1152114.
- Giraldo P, Ruiz M, Rodrı′guez-Quijano M, Benavente E. 2016. Development and validation of chloroplast DNA markers to assist Aegilops geniculata and Aegilops neglecta germplasm management. Genet Resour Crop Evol. SHORT COMMUNICATION.
- Gorenflot R, Raicu P. 1980. Cytogénétique et évolution. Paris: Masson; p. 304.
- Haider N, Nabulsi I. 2008. Identification of Aegilops L. species, and Triticum aestivum L. based on chloroplast DNA. Genet Resour Crop Evol. 55:537–549. 10.1007/s10722-007-9259-9
- Hammer K. 1980. Vorarbeiten zur Monographischen Darstellung von Wildpflanzen-Sortimenten: Aegilops L. Kulturpflanze 33–180.10.1007/BF02014641
- Hammer Ø. 2017. PAST 315. http://folk.uio.no/ohammer/past [accessed April 2017]
- Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: Paleontological Statistics sofware package.
- Hamouche Y, Amirouche N, Misset M, Amirouche A. 2010. Cytotaxonomy of autumnal flowering species of Hyacinthaceae from Algeria. Plant Syst Evol. 285:177–187.10.1007/s00606-010-0275-4
- Hatami-Maleki H, Samizadeh H, Asghari-Zakaria R, Moshtaghi N, Fazeli-Sangari E. 2010. A comparative study of chromosome morphology among some accessions of Aegilops crassa. Afr J Biotech. 9:996–1000.10.5897/AJB
- Jahier J, Chevre AM, Delourme R, Eber F, Tangay AM. 1992. Techniques de cytogénétique végétale. Editions de l’. Paris: INRA; p. 184.
- Khedim T, Amirouche N, Amirouche R. 2015. Morphological and cytotaxonomic data of Allium trichocnemis and A. seirotrichum (Amaryllidceae) endemic to Northen Algeria, compared with A. cupanii. Phytotaxa. 243 (3):247–259.doi: 10.11646/phytotaxa.243.3.3.
- Kihara H. 1946. Definieninqiand Klassification der Aegilops-Arten auf grund der genomanalyse. Seiken Ziho. 3:23–41.
- Kihara H. 1954. Considerations on the evolution and distribution of Aegilops species based on the analyzer-method. Cytologia. 19:336–357. 10.1508/cytologia.19.336
- Kilian K, Mammen K, E Millet, Sharma R, Graner A, Salamini F, Hammer K, Zkan H. 2011. chapiter 1: Aegilops. Wild crop relatives genomic and breeding resources. Berlin: Springer; p. 1–77. doi:10.1007/978-3-642-14228-4_1.
- Kimber G, Feldman M. 1987. Wild wheat An introduction. Special Report 353, College of Agriculture, University of Missouri. Columbia, SC; p. 142.
- Levan A, Fredga K, Sandberg A. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Levin DA. 2002. The role of chromosomal change in plant evolution. New York (NY): Oxford University Press; p. 226.
- Maamri F, Zermane N, Baik N, Nait Merzoug S, Bellahreche A. 2016. Karyotype studies on eight populations of Plantago albicans L.from Algeria. Caryologia. 69:102–110. doi:10.1080/00087114.2015.1109952.
- Mahjoub A, Abdellaoui R, Ben Naceur M, Ben Brahim N. 2010. Genetic diversity of Tunisian accessions of Aegilops geniculata Roth and durum wheats (Triticum durum Desf.) using RAPD markers. Acta Bot Gallica 157 (1) : 3–12. 10.1080/12538078.2010.10516184
- Maire R. 1955. Flore de l’Afrique du Nord. L Chevalier, editor. Vol. III, Paris; p. 65–69.
- Medouri A, Bellil I, Khelifi D. 2015. The genetic diversity of gliadins in Aegilops geniculata from Algeria. Czech J Genet Plant Breed. 51:9–15.
- Mirzaghaderi G, Marzangi K. 2015. IdeoKar: an ideogram constructing and karyotype analyzing software. Caryologia. 68:31–35.
- Ohta S, Nosaka Y, Yamagata H. 2015. Allopatric distributions of tetraploid and hexaploid forms of Aegilops neglecta Req. ex Bertol. Gen Res and Crop Evol. 63:193–197.
- Otto S, Whitton J. 2000. Polyploid incidence and evolution. Annu Rev Genet. 34:401–437. 10.1146/annurev.genet.34.1.401
- Paszko B. 2006. A critical review and a new proposal of karyotypes asymmetry indices. Plant Syst Evol. 258:39–48.10.1007/s00606-005-0389-2
- Peruzzi L, Altınordu F. 2014. A proposal for a multivariate quantitative approach to infer karyological relationships among taxa. Comp Cytogenet. 8(4):337–349. doi:10.3897/CompCytogen.v8i4.8564.
- Peruzzi L, Eroglu HE. 2013. Karyotype asymmetry: again, how to measure and what to measure? Comparative Cytogenetics. 7(1):1–9. doi:10.3897/CompCytogen.v7i1.4431.
- Petersen G, Seberg O, Yde M, Berthelsen K. 2006. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol Phylogenet Evol. 39(1):70–82. doi:10.1016/j.ympev.2006.01.023.
- Pignone D, Galasso I, Hammer K, Perrino P. 1994. Cytogenetic and genetic relationships between populations of Aegilops ventricosa Tausch. Euphytica. 79:81–85.10.1007/BF00023579
- Quezel P, Santa S. 1962. Nouvelle flore de l’Algérie et des régions désertiques méridionales. Tome I : Edition du. Paris: CNRS; p. 558.
- Ramsey J, Schemske DW. 2002. Neopolyploidy in flowering plants. Annu Rev Ecol Syst. 33:589–639.10.1146/annurev.ecolsys.33.010802.150437
- Rodriguez-Quijano M, Nieto-Taladriz MT, Carrillo JM. 2000. Polymorphism of high molecular weight glutenin subunits in three species of Aegilops. Genet Resour Crop Evol. 48:599–607.
- Roy RP. 1959. Genome analysis of Aegilops sharonensis. Genetica. 29:331–357.10.1007/BF01535720
- Sakamoto S, Kobayashi H. 1980. Variation and geographical distribution of cultivated plants, their wild relatives and weeds native to Turkey, Greece and Romania. Reprint from: Tani Y., Preliminary report of comparative studies on the agrico-pastoral peoples in southwestern Eurasia, II. Research Institute for the Humanistic Studies, Kyoto University, Japan pp 41-104.
- Salina EA, Lim KY, Badaeva ED, Shcherban AB, Adonina IG, Amosova AV, Samatadze TE, Vatolina TY, Zoshchuk SA, Leitch AR. 2006. Phylogenetic reconstruction of Aegilops and the evolution of Tendem repeats in the diploids and derived wheat polyploids. Genome. 49:1023–1035.10.1139/G06-050
- Sasanuma T, Chabane K, Endo TR, Valkoun J. 2004. Characterization of genetic variation and phylogenetic relationships among diploid Aegilops species by AFLP: incongruity of chloroplast and nuclear data. Theor Appl Genet. 108:612–618.10.1007/s00122-003-1485-8
- Sears ER. 1941. Chromosome pairing and fertility in hybrids and amphiploids in the Triticeae. Res Bull Hisse Agr Exp St. 337:1–120.
- Senjaninova – Korchagina M. 1932. A critical-systematical survey of the genus Aegilops L. Bull Appl Bot Genet. Plant Breed Ser II, p. 1–90.
- Soltis DE, Soltis PS. 1999. Polyploidie : recurrente formation and genome evolution. Trends Ecol Evol. 14:348–352.10.1016/S0169-5347(99)01638-9
- Stebbins GL. 1971. Chromosomal evolution in higher plants. Arnold, editor. London: Edward Arnold; p. 216.
- Stewart P. 1974. Un nouveau climagramme pour l’Algérie et son application au barrage vert. Bull Soc Hist Nat Afrique du Nord. 65:239–248.
- Sun X, Hu W, Liu S, Qian A, Hao D. 2006. Characterization of HMW glutenin subunits from Aegilops searsii and identification of a novel variant HBM glutenin subunit. Theor Appl Genet. 113(4):631–641. 10.1007/s00122-006-0327-x
- Sybenga J. 1975. The quantitative analysis of chromosomal pairing and chiasma formation based on the relative frequencies of MI configurations. VII: autotetraploids. Chromosoma. 50:211–222.
- Uslu E, Tekin Babaç M, Yılmaz A. 2012. Karyological studies on some Crocus L. taxa from Turkey. Caryologia. 65:7–10. doi:10.1080/00087114.2012.678075.
- Van Slageren MW. 1994. Wild Wheats: a monograph of Aegilops L and Amblyopyrum (Jaub & Spach) Eig (Poaceae). Wageningen Agricultural University Wageningen. TheNetherlands Papers; p. 94–97.
- Zaharieva M, Gaulin E, Havaux M, Acevedo E, Monneveux P. 2001. Drought and heat responses in the wild wheat relative Aegilops geniculata Roth: potential interest for wheat improvement. Crop Sci. 41:1321–1329.10.2135/cropsci2001.4141321x
- Zhang XY, Wang R, Dong YS. 1996. RAPD polymorphisms in Aegilops geniculata Roth. (Ae. ovata auct. non L.). Genet Resour Crop Evol. 43:429–433.
- Zhukovsky PM. 1928. Kritiko-systematischeskii obzor vydov roda Aegilops L. (Specierum generis Aegilops L. revisio critica). Trudy Prikl Bot Genet Selekc. 18:417–609 in Russian.
- Zohary D. 1966. Clonizer species in the wheat group. New York and London: Academic Press; p. 419.

