Abstract
Oligosarcus is a small genus of fish of the Characidae family and currently has 24 species widely distributed in Brazilian hydrographic systems. Cytogenetic studies were carried out in a few species of the genus, most of them from the Paraná river basin, as is the case of O. paranensis that exhibited interpopulational variations. In the present study, two populations of O. paranensis were analyzed cytogenetically in order to verify if the geographic distance between them generated some karyotypic alteration. Both populations exhibited 50 chromosomes and only two 5S rDNA sites. A marked variability in karyotype formulas, distribution of heterochromatic regions and 18S rDNA patterns were observed and their origin reflects the participation of different chromosomal rearrangements, which are considered the major events involved in the chromosome evolution of this group of fish.
Introduction
Within the order Characiformes, Characidae is the largest and most complex family, with 1126 valid species (Eschmeyer and Fong Citation2017). These fish have the widest geographical distribution in this order (Menezes et al. Citation2007), occupying nearly all freshwater environments of the American continents, from the Mexico–USA border to southern Argentina (Lucena Citation1993).
There are currently 24 valid species described for the genus Oligosarcus Günther, 1864 (Frose and Pauly Citation2017). The species of this genus are popularly known as saicanga, whitefish and dogfish (Menezes Citation1987); their habits are carnivorous, feeding on insects, crustaceans and small fish (Braga Citation1994) and their geographic distribution is restricted to South America (Menezes and Ribeiro Citation2010). The phylogenetic position of Oligosarcus has undergone several modifications. Initially, it was included in the genus Acestrorhynchus Eigenmann and Kennedy, 1903, subfamily Acestrorhynchinae (Britski et al. Citation1988). However, Buckup (Citation1998) proposed the withdrawal of the genus from this subfamily and its inclusion in the subfamily Tetragonopterinae (Characidae), being probably more related to the genus Astyanax Baird and Girard, 1984. Currently, Oligosarcus belongs to the Incertae sedis group of the family Characidae (Lima et al. Citation2003), due to its undefined or unknown phylogenetic relationships with other groups.
Cytogenetic studies in the genus Oligosarcus are restricted mostly to the species O. hepsetus Cuvier, 1829; O. jenysii Günther, 1864; O. macrolepis Steindachner, 1877; O. paranensis Menezes and Géry, 1983; O. longirostris Menezes and Géry, 1983; and O. pintoi Amaral Campos, 1945. All of them show a diploid number of 50, with interspecific and intraspecific variation in the karyotypic formulae (Table ).
Table 1. Karyotypic characteristics of the genus Oligosarcus.
Due to the variability in the karyotype structure observed in the genus Oligosarcus, the purpose of this study was to characterize and compare the karyotype of Oligosarcus paranensis from two distant locations to verify if interpopulational variations could be occurring and to discuss mechanisms involved in their karyotype evolution and possible isolation reproductive.
Material and methods
Twenty-seven individuals of Oligosarcus paranensis were analyzed: 15 specimens (10 females and five males – Museum of Zoology of the State University of Londrina [MZUEL] 16084/16085/16086/16087/16088) from the Três Bocas Stream (23°23′06.6ʺS, 51°04′35.8ʺW), belonging to the Tibagi River basin/PR and 12 individuals (four females and eight males – MZUEL 16089/16090) from the Quexada River (23°56′9.65ʺS, 51°39′26.08ʺW), Ivaí River basin/PR. The samples were collected with the permission of Instituto Brasileiro do Meio Ambiente e dos Recursos Naturais Renováveis (IBAMA), protocol number 11399-1.
Mitotic chromosomes were obtained by direct preparations of anterior kidney cells and short-term culture of solid tissues, according to Bertollo et al. (Citation1978) and Fenocchio et al. (Citation1991), respectively, and then stained with 5% Giemsa in phosphate buffer (pH 6.8). The chromosomes were organized according to Levan et al. (Citation1964) and for determination of the fundamental number (FN) considering the metacentric, submetacentric, and subtelocentric chromosomes as biarmed, and the acrocentric, as uniarmed.
Silver nitrate staining revealing active nucleolus organizer regions (Ag-NORs) was performed according to Howell and Black (Citation1980) and the distribution of constitutive heterochromatin was analyzed by Giemsa C-banding after treatments with 0.1 M HCl, Ba(OH)2 and 2XSSC (Sumner Citation1972). GC- and AT-rich segments were visualized using chromomycin A3 (CMA3) and 4-6-diamino-2-phenylindole (DAPI) fluorochrome staining (Schweizer Citation1976). Fluorescence in situ hybridization (FISH) was used for mapping 18S and 5S ribosomal DNA (rDNA) sites (Pinkel et al. Citation1986). The 18S rDNA probe from Prochilodus argenteus Agassiz 1829 (Hatanaka and Galetti Jr Citation2004) was labeled with biotin-14-dATP by nick translation using the BioNick Labeling System (Invitrogen®, Carlsbad, CA), and detected with avidin-FITC (conjugated fluorescein isothiocyanate-avidin). The 5S rDNA probe from Imparfinis schubarti (Gouveia et al. Citation2016) was labeled with digoxigenin-11-dUTP (Roche Applied Science, Mannheim, Germany) by PCR and detected with anti-digoxigenin (with rhodamine conjugate) following the manufacturer’s instructions. All images were acquired with a Leica DM 4500 B microscope (Leica Microsystem, Germany) equipped with a DFC 300FX camera and Leica IM50 4.0 software.
Results and discussion
All individuals exhibited 50 chromosomes with different formulae between populations. The individuals from the Três Bocas Stream presented 8 m+18sm+10st+14a and FN = 86 (Figure (a)) and the specimens from the Quexada River/PR, 6 m+10sm+16st+18a and FN = 82 (Figure (b)); a pair of metacentric chromosomes distinctly larger than the remainder of the chromosomal complement was evidenced in both populations, as well as in other species of Oligosarcus already analyzed (Kavalco et al. Citation2005; Centofante et al. Citation2006; Hattori et al. Citation2007). This cytotaxonomic feature has remained constant in many representatives of the family Characidae, despite its expressive karyotypic variability (Torres-Mariano and Morelli Citation2008; Paiz et al. Citation2015; Sánchez-Romero et al. Citation2015).
Figure 1. Karyotypes with conventional Giemsa staining of Oligosarcus paranensis from (a) Três Bocas Stream; and (b) Quexada River.
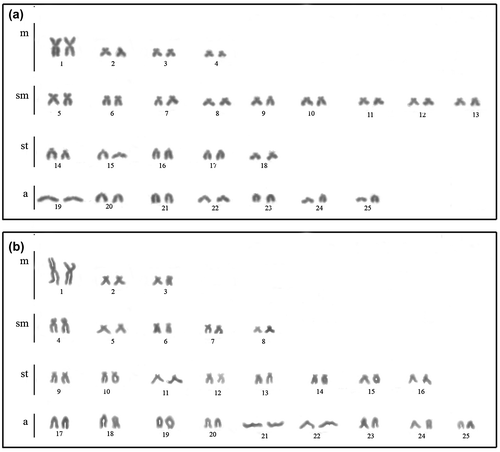
The conservation of the diploid number and considerable variability in karyotypic formulae in Oligosarcus (Table ) can be attributed to structural rearrangements, such as pericentric inversion and/or translocations, as already reported for other species of Oligosarcus (Kavalco et al. Citation2005; Barros et al. Citation2015). The two populations studied here may be in the same basin but in very distant locations and this can make genetic flow impossible and facilitate the fixation of the karyotype differences resulting from chromosomal rearrangement, as suggested by Takagui et al. (Citation2017). This reaffirms the role of pericentric inversions in the postzygotic isolation and, hence, increasing reproductive isolation and speciation (Faria and Navarro Citation2010).
The nucleolus organizer regions (AgNORs) occurred on different chromosomes in the terminal region. O. paranensis from the Três Bocas Stream showed 2–8 positive chromosomes, and the population of the Quexada River, 2–6 AgNORs (Figure (a), (e) respectively), with inter and intraindividual variation in both populations. Wide variation in location and number of sites Ag-NORs are well documented in Oligosarcus, comprising intra and interspecific variations (Table ).
Figure 2. Somatic metaphases of Oligosarcus paranensis from (a, b, c, d) Três Bocas Stream; and (e, f, g, h) Quexada River; (a, e) after silver nitrate staining; (b, f) FISH with 18S rDNA probe (red) and 5S rDNA (green); (c, g) C-banding; and (d, h) CMA3 staining. In (d), the box shows one chromosome pair with fluorescent labeling in the terminal region of the short and long arm.
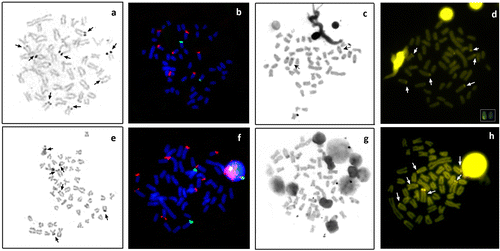
FISH with 18S rDNA probe confirmed multiple sites, all of them in terminal regions: seven chromosomes from the Três Bocas Stream population (Figure (b)) and nine chromosomes from the Quexada River (Figure (f)). The absence of 18S rDNA signal in one of the homologous chromosomes can be due to the size heteromorphism of the site, fact which makes its detection difficult by this technique. As this site is located at the chromosomes terminal area, the transference of genetic material might be occurring, from one homologous to another one, due to the proximity to the interphasic core according to the Rabl model (Cowan et al. Citation2001).
The most common in the genus is the presence of multiple sites of 18S rDNA (Table ), except for O. jenynsii (Hattori et al. Citation2007) and O. longirostris (Rubert Citation2004), which has only a pair carrier of this site. O. argentus, O. paranensis and O. solitarius are the species with more 18S rDNA sites, showing up to 11 rDNA sites (Table ). The distribution of this rDNA in Oligosarcus, according to Barros et al. (Citation2015), has been an efficient population marker.
Both populations showed one pair with 5S rDNA sites (Figure (b), (f)). Barros et al. (Citation2015) suggest that this region is very stable, as reported in other families and genera of Characiformes. Analyzing populations of O. solitarius and O. argenteus, Barros (Citation2012) suggests that this probe is not suitable for rescue of cytogenetic variability between recently isolated populations. However, among the Oligosarcus populations analyzed to date, 37.5% have more than one pair bearing this site. It is interesting to note that for O. hepsetus and O. pintoi all populations analyzed had more than one pair with 5S rDNA (Table ).
The population of the Três Bocas Stream showed size heteromorphism of this cistron (Figure (b)) and the possible causes of this structural polymorphism may be spontaneous duplication and deletions resulting from unequal crossing-over events (Gold et al. Citation1990).
The simultaneous use of 18S and 5S rDNA probes showed the lack of synteny between these sites in the two populations of Oligosarcus paranensis (Figure (b), (f)), as well as in populations of O. argenteus and O. solitarius analyzed by Barros et al. (Citation2015), contradicting the proposal of Hattori et al. (Citation2007) in O. hepsetus, O. pintoi and O. jenynsii, of syntenic organization of 18S and 5S rDNA. However, this study was not conducted to double FISH and therefore cannot show exactly the synteny of the sites.
Heterochromatin was weakly distributed in the pericentromeric region of some chromosomes and found in conspicuous blocks in the terminal regions of some chromosomes of the st-a group in both populations (Figure (c), (g)).The population from the Três Bocas River exhibited one pair with heterochromatin blocks on the short and long arm in the terminal region (Figure (c)). The heterochromatin distribution pattern in the terminal region of acrocentric chromosomes seems to be characteristic of this genus, since it was observed in all species analyzed to date (Table ).
After treatment with fluorochromes, O. paranensis presented seven CMA3+ markings all in the terminal region (Figure (d), (h)). The population from the Três Bocas River exhibited one pair with fluorescent markings on the short and long arm in the terminal region (Figure (d – box)), probably the same pair shown by C-banding. O. hepsetus analyzed by Kavalco et al. (Citation2005) also presented heterochromatin rich in GC base pairs, with this being the only work of this nature, until now, in Oligosarcus.
The different chromosomal markers used in this study enabled the characterization of the two Oligosarcus populations, confirming inter and intraspecific variability in relation to the karyotype formula and number of 18S rDNA sites. Such variations may have originated from chromosomal rearrangements, as pericentric inversions and/or translocations, which would be important events involved in the karyotype evolution of this group of fish. The distance between populations can facilitate the fixation of these variations and enabling reproductive isolation and speciation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This research was funded by grant from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ). The experiments and euthanasia of specimens were approved by the Committee of Animal Ethics (CEUA/UEL) from Universidade Estadual de Londrina.
Acknowledgments
The authors are grateful to Fernando Camargo Jerep, Department of Animal and Plant Biology (BAV), center of Biological Sciences, for identifying the species studied.
References
- Barros LC. 2012. Diferenciação vicariante recente de Oligosarcus argentus Günther, 1864 e Oligosarcus solitarius Menezes, 1987 nas bacias do rio Doce e São Francisco, Minas Gerais, Brasil [ dissertation]. Viçosa (MG): Universidade Federal de Viçosa.
- Barros LC, Santos U, Cioffi MB, Dergam JA. 2015. Evolutionary divergence among Oligosarcus spp. (Ostariophysi, Characidae) from the Sao Francisco and Doce River basins: Oligosarcus solitarius Menezes, 1987 shows the highest rates of chromosomal evolution in the neotropical region. Zebrafish. 12:102–110.10.1089/zeb.2014.1030
- Bertollo LAC, Takahashi CS, Moreira-Filho O. 1978. Cytotaxonomic considerations on Hoplias lacerdae (Pisces Erythrinidae). Braz J Genet. 1:103–120.
- Braga L. 1994. Los Characidae de Argentina de las subfamilias Cynopotaminae e Acestrorhynchinae. Fauna de água Dulce de La Republica Argentina, PROFADU (CONICET). 40(6):5–45.
- Britski H, Sato Y, Rosa ABS. 1988. Manual de Identificação de peixes da região de Três Marias: com chaves de identificação para os peixes da bacia do São Francisco. 3rd ed. Brasília: CODEVASF.
- Buckup PA. 1998. Relationships of the Characidiinae and phylogeny of characiform fishes Teleostei: Ostariophisy). In: Malabarba LR, Reis RE, Vari RP, Lucena ZM, Lucena CA, editors. Phylogeny and classification of neotropical fishes. Porto Alegre (RS): Edipucrs; p. 123–144.
- Centofante L, Bertollo LAC, Moreira-Filho O. 2006. Chromosomal differentiation between populations of Oligosarcus hepsetus (Teleostei, Characidae) from small tributaries at opposite margins of the Paraíba do Sul River (Brazil). Braz Arch Biol Technol. 49:981–987.10.1590/S1516-89132006000700016
- Cowan CR, Carlton PM, Cande WZ. 2001. The polar arrangement of telomeres in interphase and meiosis. Rabl organization and the bouquet. Plant Physiol. 125:532–538.10.1104/pp.125.2.532
- Cunha EB, Pegoraro JL, Margarido VP. 2001. Análise Citogenética comparativa em duas espécies de Oligosarcus (Pisces, Characidae, Acestrorhynchinae). Poster session presented at: 47º Congresso Nacional de Genética, Águas de Lindóia, Brazil.
- Eschmeyer WN, Fong JD. 2017. Catalogue of fishes. Electronic version. [ accessed Jun 9]. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp.
- Falcão JN, Bertollo LAC. 1985. Chromosome characterization in Acestrorhynchinae and Cynopotaminae (Pisces, Characidae). J Fish Biol. 27:603–610.10.1111/jfb.1985.27.issue-5
- Faria R, Navarro A. 2010. Chromosomal speciation revisited: rearranging theory with pieces of evidence. Trends Ecol Evol. 25:660–669.10.1016/j.tree.2010.07.008
- Fenocchio AS, Venere PC, César ACG, Dias AL, Bertollo LAC. 1991. Short term culture from solid tissues of fishes. Caryologia. 44:161–166.10.1080/00087114.1991.10797181
- Frose R, Pauly D. 2017. Catalogue of life: FishBase. Electronic version. [accessed 2017 Jun 9. www.fishbase.org.version.
- Gold JR, Li C, Shipley NS, Powers PK. 1990. Improved methods for working with chromosomes with a review of metaphase chromosome banding. J Fish Biol. 37:563–575.
- Gouveia JG, Wolf IR, Moraes-Manécolo VPO, Bardella VB, Ferracin LM, Giuliano-Caetano L, da Rosa R, Dias AL. 2016. Isolation and characterization of 5S rDNA sequences in catfishes genome (Heptapteridae and Pseudopimelodidae): perspectives for rDNA studies in fish by C0t method. Cytotechnology. 68:2711–2720.10.1007/s10616-016-9996-8
- Hatanaka T, Galetti PM Jr. 2004. Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae). Genetica. 122:239–244.10.1007/s10709-004-2039-y
- Hattori RS, Daniel-Silva MFZ, Almeida-Toledo LF. 2007. Karyotype characterization and gene mapping of 5S and 18S rDNA in three species of Oligosarcus (Teleostei: Characidae). Caryologia. 60:372–378.10.1080/00087114.2007.10797961
- Howell WM, Black DA. 1980. Controlled silver staining of Nucleolus Organizer Regions with a protective colloidal developer: a 1-step method. Experientia. 36:1014–1015.
- Kavalco KF, Pazza R, Bertollo LAC, Moreira-Filho O. 2005. Molecular cytogenetics of Oligosarcus hepsetus (Teleostei, Characiformes) from two Brazilian locations. Genetica. 124:85–91.10.1007/s10709-005-0176-6
- Levan A, Fregda K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Lima FCT, Malabarba LR, Buckup PA, Silva JFP, Vari RP, Harold A, Benine R, Oyakawa OT, Pavanelli CS, Menezes NA, et al. 2003. Genera Incertae Sedis in Characidae. In: Reis RE, Kullander SO, Ferraris CJ Jr, editors. Check list of the freshwater fishes of South and Central America. Porto Alegre: Edipucrs; p. 106–169.
- Lucena CAS. 1993. Estudos filogenéticos da família Characidae com uma discussão dos grupos naturais propostos (Teleostei, Ostariophysi, Characiformes) [Doctoral thesis]. São Paulo (SP): Universidade de São Paulo.
- Martinez ERM, Oliveira C, Júlio HF Jr. 2004. Cytogenetic analysis of species of the genera Acestrorhynchus, Oligosarcus and Rhaphiodon (Teleostei: Characiformes). Caryologia. 57:294–299.10.1080/00087114.2004.10589408
- Menezes NA. 1987. Três novas espécies de Oligosarcus Günther, 1864 e redefinição taxonômica das demais espécies do gênero (Osteichthyes, Teleostei, Characidae). Boll Zool. 11:1–39.
- Menezes NA, Ribeiro AC. 2010. Oligosarcus jacuiensis (Characiformes: Characidae), a new species from the Uruguay and Jacuí River basins, southern Brazil. Neotrop Ichthyol. 8:649–653.10.1590/S1679-62252010000300010
- Menezes NA, Weitzman SH, Oyakawa OT, Lima FCT, Castro RMC, Weitzman MJ. 2007. Peixes de água doce da Mata Atlântica – Lista preliminar das espécies e comentários sobre conservação de peixes de água doce neotropicais. São Paulo (SP): Museu de Zoologia Universidade de São Paulo.
- Paiz LM, Baumgärtner L, Graça WJ, Margarido VP. 2015. Basic cytogenetics and physical mapping of ribosomal genes in four Astyanax species (Characiformes, Characidae) collected in Middle Paraná River, Iguassu National Park: considerations on taxonomy and systematics of the genus. Comp Cytogenet. 9:51–65.
- Pinkel D, Straume T, Gray JW. 1986. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci. 83:2934–2938.10.1073/pnas.83.9.2934
- Rubert M. 2004. Análises citogenéticos em três espécies do gênero Oligosarcus [Monograph]. Cascavel (PR): Universidade Estadual do Oeste do Paraná.
- Rubert M, Margarido VP. 2007. Cytogenetic studies in three species of the genus Oligosarcus. Braz Arch Biol Technol. 50:127–135.10.1590/S1516-89132007000100015
- Sánchez-Romero O, Abad CQ, Cordero PQ, de Sene VF, Nirchio M, Oliveira C. 2015. First description of the karyotype and localization of major and minor ribosomal genes in Rhoadsia altipinna Fowler, 1911 (Characiformes, Characidae) from Ecuador. Comp Cytogenet. 9(2):271–280.
- Schweizer D. 1976. Reverse fluorescent chromosome banding with Chromomycin and DAPI. Chromosoma. 92:143–148.
- Sumner AT. 1972. A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 75:304–306.10.1016/0014-4827(72)90558-7
- Takagui FH, da Rosa R, Shibatta OA, Giuliano-Caetano L. 2017. Chromosomal similarity between two species of Apteronotus albifrons complex (Apteronotidae–Gymnotiformes) implications in cytotaxonomy and karyotypic evolution. Caryologia. 70:147–150.10.1080/00087114.2017.1306385
- Torres-Mariano AR, Morelli S. 2008. B chromosomes in a population of Astyanax eigenmanniorum (Characiformes, Characidae) from the Araguari River basin (Uberlândia, MG, Brazil). Genet Mol Biol. 31:246–249.10.1590/S1415-47572008000200015
