ABSTRACT
Nuclear genome size (C-value) was determined by flow cytometry for 23 populations of 15 Crepis L. (Asteraceae) taxa originating from Turkey. Genome size of seven taxa are reported for the first time as well as two new chromosome counts. The 1C-value of the studied taxa ranges from 1.35 pg to 7.06 pg, which represents more than fivefold variation. There is a positive correlation between genome size and altitude. Genome size is not correlated with the chromosome number and sectional division of the studied taxa. Systematic and ecological significance of genome size are discussed within the framework of the results obtained in this study.
Introduction
The genus Crepis L. belongs to the tribe Cichorieae of the Asteraceae family (Compositae) and comprises over 200 species (Bremer Citation1994) distributed throughout the northern hemisphere and Africa (Enke Citation2009). It includes annual, biennial and perennial herbs. Crepis has been classified into 27 sections by Babcock (Citation1947a, Citation1947b) based on morphology, geographical distribution, chromosome number and karyotype composition. However, Enke and Gemeinholzer (Citation2008) showed that the infrageneric classification of Babcock (Citation1947a, Citation1947b) was in many cases inconsistent with molecular clades and Enke (Citation2009) indicated that Babcock’s sections do not represent natural groups.
Crepis is thought to have originated in the Central Asia Altai/Tien Shan region (Babcock Citation1947a), and to have spread from there towards Europe and the Mediterranean (Enke et al. Citation2011). The geographical situation of Turkey presupposes active formation processes in the genus Crepis in particular, as Asia Minor has been the meeting point of migrating the genomes from Asia to the Mediterranean and Europe. The genus includes ca. 40 taxa in Turkey, of which eight are endemic.
Genome size (C-value) is an important biodiversity character, which was proved to be related to biological traits, such as life cycle, life growth form and ecology. Its availability is thus advancing research in many areas, including molecular biology, systematics, ecology and population biology (Leitch and Bennett Citation2007; Bennet and Leitch Citation2011; Garcia et al. Citation2013). Genome size can be dramatically variable among different species (Bennett and Leitch Citation2005), but it is usually constant within species or subspecies and variation in this character can, therefore, be viewed as an indicator of taxonomic heterogeneity and/or ecological parameters (Loureiro et al. Citation2010).
The systematic and ecological significance of the C-value in Crepis have been noted by previous studies (Dimitrova et al. Citation1999; Dimitrova and Greilhuber Citation2000; Enke et al. Citation2011, Citation2015; Inceer et al. Citation2016). However, the C-values of most Crepis species are still unknown, and the C-value data are especially very scarce for the Crepis taxa from Turkey. The aim of the present study is to characterize the genome of 15 Crepis taxa (23 populations) growing in Turkey by genome size assessment together with chromosome counts, and to detect any genome size variability among populations of the taxa.
Material and methods
Plant material
The plant materials of the Crepis taxa investigated in this study were collected from natural populations in Turkey. Taxonomic identification and verification of the specimens were performed following the keys of Crepis taxonomy of Babcock (Citation1947b) and Lamond (Citation1975) as well as Ekim (Citation2012). Vouchers are deposited in the herbarium at the Karadeniz Technical University, Department of Biology (KTUB). The names of the investigated taxa, accession numbers and collection information are given in .
Table 1. Localities and voucher numbers of Crepis taxa investigated.
Germination of achenes and chromosome counts
Mature achenes obtained from five different individuals per population were germinated in Petri dishes at 22–23°C. The germinated seedling with well-developed root tips of about 1 cm length were pre-treated with 0.05% colchicine solution for 2–5 h at room temperature. They were fixed in absolute ethanol-glacial acetic acid (3:1) for at least 24 h at 4°C, hydrolysed in 1M HCl at 60°C for 10 min and then rinsed in deionized water for 2–3 min. Staining was carried out in 1% aqueous lacto-propionic orcein for 12–18 h at room temperature, squashes were made in 45% acetic acid, and the preparations were mounted in Entellan (Inceer et al. Citation2016). Permanent slides were observed with Leica DM 4000B microscope (Wetzlar, Germany) at a magnification of 1000 ×. Best metaphase plates were photographed using a Leica DFC 490 digital camera. Chromosome counts were carried out on five metaphase plates from five individuals per population.
Flow cytometric measurements
For flow cytometric measurements, plants were grown from achenes obtained from five different individuals in a plant grow room. Nuclear DNA amounts were determined according to Doležel et al. (Citation2007). Small pieces of young leaf tissue of both analysed species and reference standard plant were transferred to a glass Petri dish. The tissue was chopped in 500 µl Otto I solution (0.1 M citric acid, 0.5% v/v Tween 20; Otto Citation1990). The crude suspension of nuclei was filtered through a 50-µm nylon mesh. Nuclei were then pelleted (500 g, 3 min) and resuspended in 400 µl of Otto I. After 30 min incubation at room temperature, 800 µl of Otto II solution (0.4 M Na2HPO4; Otto Citation1990), supplemented with 50 µg ml–1 RNase and 50 µg ml–1 propidium iodide (PI), were added. Samples were then analysed using Partec PAS flow cytometer (Partec GmbH, Münster, Germany) equipped with a 488-nm argon laser. Due to differences in genome size of analysed species, two reference standards were used for flow cytometric analysis. Pea (Pisum sativum ‘Ctirad’; 2C = 9.09 pg DNA, Doležel et al. Citation1998) served as an internal standard for DNA content estimation in Crepis bupleurifolia, C. pulchra and C. smyrnaea and maize (Zea mays ‘CE-777’; 2C = 5.43 pg DNA, Lysak and Dolezel Citation1998) was used for all remaining taxa. The gain of the instrument was adjusted so that the peak representing G1 (2C) nuclei of the standard appeared approximately on channel 100 on a histogram of relative fluorescence intensity when using a 512-channel scale.
Three plants were measured per accession and each individual was analysed three times on three different days. At least 5000 nuclei per sample were analysed and the absolute DNA content was then calculated from mean values of G1 peaks following the formula:
Statistical analysis
The correlation analysis was carried out to evaluate the relationships between genome size and other factors, such as chromosome number, altitude and section. The statistical analysis was performed with using SPSS version 17 (SPSS Inc, Chicago, USA).
Results
The results obtained from flow cytometric analysis of propidium iodide-stained nuclei are summarized in . Chromosome numbers of the studied taxa are 2n = 2x = 8, 10 and 12 with three different basic chromosome numbers (x = 4, 5, 6, , and 1C-value varies from 1.35 pg to 7.06 pg.
Table 2. Genome size (C-value), somatic chromosome number (2n) and life cycle for the studied Crepis taxa.
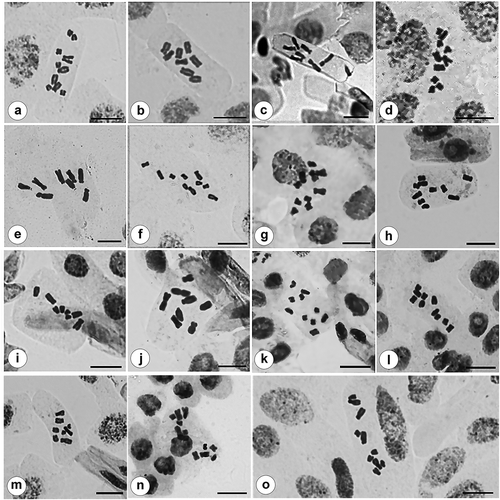
Figure 1. Somatic metaphases in Crepis. (a) C. alpina (2n = 10); (b) C. aspera (2n = 8); (c) C. bupleurifolia (2n = 8); (d) C. commutata (2n = 10, Inceer 844); (e) C. dioscoridis (2n = 8); (f) C. foetida subsp. foetida (2n = 10, Aksu 133); (g) C. foetida subsp. glandulosa (2n = 10); (h) C. foetida subsp. rhoeadifolia (2n = 10, Inceer 955); (i) C. micrantha (2n = 8); (j) C. pulchra (2n = 8, Inceer 796); (k) C. sancta subsp. nemausensis (2n = 10, Inceer 940); (l) C. sancta subsp. sancta (2n = 10); (m) C. setosa subsp. setosa (2n = 8); (n) C. smyrnaea (2n = 12); (o) C. syriaca (2n = 10). Scale bars = 10 µm.
The 1C-value within the annual taxa varies from 1.35 pg in C. commutata to 5.72 pg in C. pulchra. The 1C-value within rhizomatous perennial species ranges from 2.64 pg in C. symrnaea to 7.06 pg in C. bupleurifolia. The mean 1C-value within the annual taxa studied is 2.39 pg whereas the mean 1C-value within rhizomatous perennial ones is 4.85 pg. Within the sampled taxa, both very small and small genomes are present in only C. commutata. Intermediate genomes are found in C. bupleurifolia, C. dioscoridis and C. pulchra whereas other taxa have small genomes ().
Within the sampled taxa, the 1C-value is not correlated with chromosome number (Spearman’s rho, r = – 0.306, p = 0.156). A positive correlation is found between 1C-value and altitude (Pearson’s r = 0.45, p < 0.05) in the studied taxa. There is no correlation between 1C-value and sectional division of the studied taxa (Spearman’s r = 0.112, p = 0.609).
Among the studied taxa, there are some differences in 1C-value between their populations of some Crepis taxa (). The intraspecific variation in 1C-value ranges from 9.8% in C. sancta up to 34.8% in C. commutata. At intrasubspecific level, the highest variation in 1C-value occurs in C. foetida subsp. rhoeadifolia (17.4%).
Table 3. 1C-value and intraspecific/intrasubspecific variability of some populations of Crepis.
Discussion
This is the first comprehensive study on nuclear genome size of the Turkish Crepis taxa, and the C-value of seven Crepis taxa, which are C. aspera, C. bupleurifolia, C. foetida subsp. glandulosa, C. micrantha, C. sancta subsp. nemausensis, C. symrnaea and C. syriaca are reported here for the first time as well as two new chromosome counts (C. bupleurifolia and C. symrnaea). More than fivefold variation in genome size is found in the studied taxa (). The present results are in agreement with previous reports in diploid Crepis species (Jones and Brown Citation1976; Dimitrova and Greilhuber Citation2000; Enke et al. Citation2011). Similar results are also reported in other members of Asteraceae (Vallès et al. Citation2013, Citation2017).
According to Leitch et al. (Citation1998) and Soltis et al. (Citation2003), genome sizes can be assigned to a series of distinct categories: very small (1C ≤ 1.4 pg), small (1.4 < 1C ≤ 3.5 pg), intermediate 3.5 < 1C < 14 pg), large (14 ≤ 1C < 35 pg) and very large (1C ≥ 35 pg). Most Asteraceae genomes (57.23%) are considered to be very small or small (1C ≤ 3.5 pg), and only 1.78% have 1C-values equal or larger than 14 pg (Vallès et al. Citation2013). So far, based on 1C-value, very small, small and intermediate genomes have been reported in the genus Crepis by a number of authors (Bennett and Smith Citation1976; Bennett et al. Citation1982; Dimitrova et al. Citation1999; Dimitrova and Greilhuber Citation2000: Enke et al. Citation2011, Citation2015; Inceer et al. Citation2016). Our results show that the studied taxa have very small, small and intermediate genomes. Present and previous studies indicate that Crepis is quite heterogenous in terms of genome size.
It is well known that, within a genus, an increase in genome size implies a longer cell cycle, thus annual plants have usually smaller genome sizes than perennial ones (Torrell and Vallès Citation2001; Garcia et al. Citation2005). Enke et al. (Citation2011) noted that the annual species of Crepis have very small, small and, in rare cases, intermediate 1C-values whereas perennials are usually more variable in genome size. Our findings show that small and intermediate 1C-values are found in both annuals and rhizomatous perennials. In addition, very small 1C-values are present in only two populations of C. commutata, which is a Mediterranean annual species.
The 1C-values of C. alpina, C. commutata, C. dioscoridis, C. foetida subsp. foetida, C. foetida subsp. rhoeadifolia, C. pulchra, C. sancta subsp. sancta and C. setosa subsp. setosa were reported in previous studies (Bennett and Smith Citation1976; Bennett et al. Citation1982; Dimitrova et al. Citation1999; Dimitrova and Greilhuber Citation2000). However, there are some differences in the 1C-values between present and previous reports of these taxa. The differences in the 1C-values might be due to the use of different types of flow cytometers and different internal standards as well as geographical or environmental factors. Similar results are reported in Arabidopsis (Doležel et al. Citation1998), Linum (Durrant and Jones Citation1971) and Artemisia (Torrell and Vallès Citation2001).
There is a relationship between selfing breeding system and annual life history, and annual selfing species have smaller genomes than out-crossing species (Albach and Greilhuber Citation2004; Enke et al. Citation2011). Some species of Crepis, e.g. C. pulchra and C. alpina, are known to be self-compatible (Babcock Citation1947a). In addition, only two species, C. foetida and C. sancta, are known with no or low self-compatibility (Babcock Citation1947a; Enke et al. Citation2011). Our results show that all annual species included in this study have very small and small 1C-values, except for C. pulchra, with an intermediate 1C-value. The present results are in agreement with the previous report in annual Crepis species (Enke et al. Citation2011).
Rayburn and Auger (Citation1990) reported an increase in genome size as an adaptation to high altitudes. Within Asteraceae, an increase in genome size has been detected in some genera, such as Hypochaeris (Cerbah et al. Citation1999), Sonchus (Suda et al. Citation2003) and Centaurea (Bancheva and Greilhuber Citation2006). However, some authors have shown a negative correlation between genome size and altitude in some genera of Asteraceae, such as Tripleurospermum (Garcia et al. Citation2005), Argyranthemum and Microseris (Suda et al. Citation2005). Our results indicate that 1C-value in the studied taxa have significantly increased in higher altitudes. The high amount of nuclear DNA increasing proportion of repetitive or non-coding sequences might influence climatic adaptation to higher altitudes of the Crepis species.
Enke et al. (Citation2011) reported that there was no significant correlation between genome size and chromosome number within Crepis. The present results are in agreement with this previous report. Our findings indicate that decrease in genome sizes is not accompanied by changes in the number of chromosomes in the studied taxa.
Despite the fact that genome size is constant within a species, it is obvious that a certain degree of genuine intraspecific variation is always possible (Greilhuber Citation1998; Garcia et al. Citation2005; Jaume et al. Citation2009; Šmarda and Bureš Citation2010; Trávníček et al. Citation2013). It is known that the differences in genome size are largely caused by different amounts of non-coding repetitive DNA, which is composed of transposable elements, satellite DNA, introns and pseudogenes as well as chromosomal heterogeneity or hybridization (Bennett and Leitch Citation2005; Suda et al. Citation2007; Dušková et al. Citation2010; Loureiro et al. Citation2010; Šmarda and Bureš Citation2010). Dimitrova and Greilhuber (Citation2000) reported intraspecific variation in genome size in some species of Crepis from Bulgarian populations. Our results show that there are some differences in genome size between the populations of the same taxon without changing chromosome number, as in C. commutata, C. foetida subsp. rhoeadifolia, C. pulchra and C. sancta subsp. nemausensis (). However, we did not detect any conspicuous morphological differentiation, or any sign of hybridization among and within populations of these taxa. Present results show that the analysed accessions of these taxa were taxonomically homogenous. The genome size variation may represent physiological and ecological differentiation within these taxa.
Ekim (Citation2012) reported two subspecies – subspp. foetida and rhoeadifolia – within C. foetida in the Turkish flora and he noted that the occurrence of C. foetida subsp. glandulosa in Turkey has required confirmation. The difference in genome size as well as morphological characters using delimitation of subspecies glandulosa from other subspecies – subspp. foetida and rhoeadifolia – confirm the presence of C. foetida subsp. glandulosa in Turkey.
According to molecular phylogenetic analysis of Crepis (Enke and Gemeinholzer Citation2008; Enke Citation2009), C. smyrnaea is very closely related to C. lapsanoides, from which it is easily distinguished by the much smaller flower heads, the glabrous receptacle, very much smaller florets, shorter achenes and pappus, and, at least in typical forms, the tawny pappus (Babcock Citation1947b). Crepis smyrnaea is a diploid species with 2n = 2x = 12 chromosomes as in C. lapsanoides (2n = 12; Babcock Citation1947a, Citation1947b), but the 1C-value differs in these homoploid species. According to genome classes of Leitch et al. (Citation1998) and Soltis et al. (Citation2003), C. smyrnaea has small genome (1C: 2.64 pg) whereas C. lapsanoides has intermediate genome (1C: 5.6 pg; Bennett and Smith Citation1976). We suggest that genome size can be used as taxonomic marker for delimitation of these homoploid species.
Babcock (Citation1947b) suggested that the sectional classification into 27 sections reflected phylogenetic relationships within Crepis, but recent phylogenetic analysis of the genus based on the molecular markers ITS and matK established 23 internal clades were not all concordant with Babcock’s earlier systematic classification (Enke and Gemeinholzer Citation2008; Enke Citation2009; Enke et al. Citation2015). According to Enke et al. (Citation2011), genome size is not correlated to Babcock’s (Citation1947b) sectional classification within Crepis. The taxa analysed in the present study represent eight of Babcock’s sections (1947b), and similarly to findings of Enke et al. (Citation2011), we found no correlation between genome size and sectional classification in the studied taxa. However, genome size of most Crepis species is still unknown. Further studies on genome size of Crepis species might shed more light on the observed trend.
Acknowledgements
The authors thank Dr Jan Bartoš and two anonymous reviewers for providing helpful comments and suggestions that improved the manuscript. The corresponding author would like to thank Council of Higher Education, Turkey, for the grant in Centre of Plant Structural and Functional Genomics of Institute of Experimental Botany AS CR.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Albach DC, Greilhuber J. 2004. Genome size variation and evolution in Veronica. Ann Bot. 94:897–911.
- Babcock EB. 1947a. The genus Crepis, part one: the taxonomy, phylogeny, distribution and evolution of Crepis. Berkeley and Los Angeles: University of California Press.
- Babcock EB. 1947b. The genus Crepis, part two: systematic treatment. Berkeley and Los Angeles: University of California Press.
- Bancheva S, Greilhuber J. 2006. Genome size in Bulgarian Centaurea s.l. (Asteraceae). Plant Syst Evol. 257:95–117.
- Bennett MD, Leitch IJ. 2005. Genome size evolution in plants. In: Gregory T, editor. The evolution of the genome. Amsterdam: Elsevier Academic Press.
- Bennett MD, Leitch IJ. 2011. Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot. 107:467–590.
- Bennett MD, Smith JB. 1976. Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci. 274:227–274.
- Bennett MD, Smith JB, Heslop-Harrison JS. 1982. Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci. 216:179–199.
- Bremer K. 1994. Asteraceae, cladistics and classification. Portland (OR): Timber Press.
- Cerbah M, Coulaud J, Brown SC, Siljak-Yakovlev S. 1999. Evolutionary DNA variation in the genus Hypochaeris. Heredity. 82:261–266.
- Dimitrova D, Ebert I, Greilhuber J, Kozhuharov S. 1999. Karyotype constancy and genome size variation in Bulgarian Crepis foetida s.l. (Asteraceae). Plant Syst Evol. 217:245–257.
- Dimitrova D, Greilhuber J. 2000. Karyotype and DNA-content evolution in ten species of Crepis (Asteraceae) distributed in Bulgaria. Bot J Linn Soc. 132:281–297.
- Doležel J, Bartos J, Voglmayr H, Greilhuber J. 2003. Nuclear DNA content and genome size of trout and human. Cytometry. 51A:127–128.
- Doležel J, Greilhuber J, Lucretti S, Meister A, Lysák MA, Nardi L, Obermayer R. 1998. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot. 82:17–26.
- Doležel J, Greilhuber J, Suda J. 2007. Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc. 2:2233–2244.
- Durrant A, Jones TWA. 1971. Reversion of induced changes in amount of nuclear DNA in Linum. Heredity. 27:431–439.
- Dušková E, Kolár F, Sklenář P, Rauchova J, Kubešová M, Fér T, Suda J, Marhold K. 2010. Genome size correlates with growth form, habitat and phylogeny in the Andean genus Lasiocephalus (Asteraceae). Preslia. 82:127–148.
- Ekim T. 2012. Crepis L. In: Guner A, Aslan S, Ekim T, Vural M, Babac MT, editors. Türkiye Bitkileri Listesi (Damarli Bitkiler). Istanbul: Nezahat Gokyigit Botanik Bahcesi ve Flora Arastirmalari Dernegi Yayini; p. 150–154.
- Enke N. 2009. Contributions towards a revised infrageneric classification of Crepis (Cichorieae, Compositae). Willdenowia. 39:229–245.
- Enke N, Fuchs J, Gemeinholzer B. 2011. Shrinking genomes? Evidence from genome size variation in Crepis (Compositae). Plant Biol. 13:185–193.
- Enke N, Gemeinholzer B. 2008. Babcock revisited: new insights into generic delimitation and character evolution in Crepis L. (Compositae: Cichorieae) from ITS and matK sequence data. Taxon. 57:756–768.
- Enke N, Kunze R, Pustahija F, Glöckner G, Zimmermann J, Oberländer J, Kamari G, Siljak-Yakovlev S. 2015. Genome size shifts: karyotype evolution in Crepis section Neglectoides (Asteraceae). Plant Biol. 17:775–786.
- Garcia S, Hidalgo O, Jakovljević I, Siljak-Yakovlev S, Vigo J, Garnatje T, Vallès J. 2013. New data on genome size in 128 Asteraceae species and subspecies, with first assessments for 39 genera, three tribes and two subfamilies. Plant Biol. 147:1219–1227.
- Garcia S, Inceer H, Garnatje T, Vallès J. 2005. Genome size variation in some representatives of the genus Tripleurospermum. Biol Plantarum. 49:381–387.
- Greilhuber J. 1998. Intraspecific variation in genome size in angiosperms: a critical reassessment. Ann Bot. 82:27–35.
- Inceer H, Aksu Kalmuk N, Imamoglu KV, Duman O, Hayirlioglu-Ayaz S, Arslan G. 2016. Micromorphological, anatomical and cytogenetical studies in endemic Crepis macropus Boiss. & Heldr. (Asteraceae) from Turkey. Acta Bot Croat. 75:173–178.
- Jaume P, Garcia S, Garnatje T, Vallès J. 2009. Changes in genome size in a fragmented distribution area: the case of Artemisia crithmifolia L. (Asteraceae, Anthemideae). Caryologia. 62(2):152–160.
- Jones RN, Brown LM. 1976. Chromosome evolution and DNA variation in Crepis. Heredity. 36:91–104.
- Lamond JM. 1975. Crepis L. In: Davis PH, editor Flora of Turkey and the East Aegean Islands. Vol. 5. Edinburgh: University Press; p. 814–841.
- Leitch IJ, Bennett MD. 2007. Genome size and its uses: the impact of flow cytometry. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Weinheim: Wiley-VCH; p. 153–176.
- Leitch IJ, Chase MW, Bennett MD. 1998. Phylogenetic analysis of C-DNA values provides evidence for a small ancestral genome size in flowering plants. Ann Bot. 82:85–94.
- Loureiro J, Trávníček P, Rauchová J, Urfus T, Vít P, Štech P, Castro S, Suda J. 2010. The use of flow cytometry in the biosystematics, ecology and population biology of homoploid plants. Preslia. 82:3–21.
- Lysak MA, Dolezel J. 1998. Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia. 51:123–132.
- Otto FJ. 1990. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Darzynkiewickz Z, Crissman HA, editors. Methods in cell biology. San Diego (CA): Academic Press; p. 105–110.
- Rayburn AL, Auger JA. 1990. Genome size variation in Zea mays ssp. mays adapted to different altitudes. Theor Appl Genet. 79:470–474.
- Šmarda P, Bureš P. 2010. Understanding intraspecific variation in genome size in plants. Preslia. 82:41–61.
- Soltis DE, Soltis PS, Bennett MD, Leitch IJ. 2003. Evolution of genome size in the angiosperms. Am J Bot. 90:1596–1603.
- Suda J, Krahulcová A, Trávníček P, Rosenbaumová R, Peckert T, Krahulec F. 2007. Genome size variation and species relationships in Hieracium sub-genus Pilosella (Asteraceae) as inferred by flow cytometry. Ann Bot. 100:1323–1335.
- Suda J, Kyncl T, Freiová R. 2003. Nuclear DNA amount in Macaronesian angiosperms. Ann Bot. 92(1):153–164.
- Suda J, Kyncl T, Jarolímová V. 2005. Genome size variation in Macaronesian angiosperms: forty percent of the Canarian endemic flora completed. Plant Syst Evol. 252(3–4):215–238.
- Torrell M, Vallès J. 2001. Genome size in 21 Artemisia L. species (Asteraceae, Anthemideae): systematic, evolutionary, and ecological implications. Genome. 44:231–238.
- Trávníček P, Kirschner J, Chudáčková H, Rooks F, Štěpánek J. 2013. Substantial genome size variation in Taraxacum stenocephalum (Asteraceae, Lactuceae). Folia Geobot. 48:271–284.
- Vallès J, Canela MÁ, Garcia S, Hidalgo O, Pellicer J, Sánchez-Jiménez I, Siljak-Yakovlev S, Vitales D, Garnatje T. 2013. Genome size variation and evolution in the family Asteraceae. Caryologia. 66:221–235.
- Vallès J, Malik S, Gomez M, Siljak-Yakovlev S. 2017. Contribution to knowledge about nuclear DNA amounts in the family Asteraceae: first assessments in one genus and 12 species, with chromosome counts for three taxa. Bot Serb. 41:213–219.
