ABSTRACT
This study examined the detailed chromosome measurements and karyotype asymmetries of seven taxa in the genus Vicia. The taxa are V. articulata, V. cassubica, V. villosa subsp. villosa in sect. Cracca, V. noeana var. noeana, V. sativa subsp. sativa, V. peregrina in sect. Vicia and V. caesarea in sect. Ervum. V. cassubica, V. noeana var. noeana, V. sativa subsp. sativa, V. caesarea have 2n = 12 chromosomes. V. articulata, V. villosa subsp. villosa, V. peregrina have 2n = 14 chromosomes in somatic cells. Total chromosome lengths range between 2.93–4.99 µm in V. articulata, 2.09–4.73 µm in V. cassubica, 1.86–3.36 µm in V. villosa subsp. villosa, 4.23–6.05 µm in V. noeana var. noeana, 2.07–3.72 µm in V. sativa subsp. sativa, 4.32–7.21 µm in V. peregrina and 2.39–5.78 µm in V. caesarea. The detailed chromosome measurements, relative lengths, centromeric indexes and karyotype asymmetries are also given. V. articulata is the most symmetrical karyotype, while V. villosa subsp. villosa is the most asymmetrical karyotype in intrachromosomal asymmetry including parameters of MCA, AsK, TF, Syi, A1, and A. However, the asymmetrical karyotypes are different in interchromosomal asymmetries. While V. noeana var. noeana is the most symmetrical karyotype in CVCL, Rec, and A2. V. caesarea is the most asymmetrical karyotype in only CVCL and A2. Unlike all parameters, V. cassubica is the most asymmetrical karyotype in Rec value. The scatter diagrams are given between MCA–CVCL and Syi–Rec.
Introduction
The family Fabaceae (Leguminosae) is the third largest family of flowering plants and contains more than 700 genera and 19,000 species (Lewis et al. Citation2005; Kahraman et al. Citation2013). The family consists of two subfamilies (Papilionoideae and Mimosoideae) whose members are monophyletic and one subfamily (Caesalpinioideae) of which each member is paraphyletic. The largest subfamily Papilionoideae contains more than 13,800 species, including important food crops. The genus Astragalus L., Dalea L., Erythrina L., Lupinus L., Lathyrus L., Robinia L., Vicia L. and Onobrychis Mill. belong to this subfamily (Miller et al. Citation2011; Kahraman et al. Citation2013; Tanaomi et al. Citation2016).
The genus Vicia is placed in the tribe Fabeae in the subfamily Papilionoideae. It contains about 180–200 species, spread throughout the temperate regions of Europe, Asia, North and South America (Kupicha Citation1976; Jaaska Citation2005). The highest specific diversity of Vicia is found in Turkey and northwest Asia (Maxted and Hawkes Citation1997). In Maxted (Citation1993), the subgenus Vicia has been divided into nine sections based on the phenetic basis. The sections are Atossa (Alef.) Asch. & Graebner, Microcarinae Maxted, Hypechusa (Alef.) Asch. & Graebner, Peregrinae Kupicha, Wiggersia (Alef.) Maxted, Vicia, Narbonensis (Radzhi) Maxted, Bithynicae (B. Fedtsch. ex Radzhi) Maxted and Faba (Mill.) Ledeb. (Kahraman et al. Citation2013).
The origin center of the subgenus Vicia is a restricted area in central northeast Mediterranean including Iraq, Iran, Turkey, Syria and the former Soviet Union (Kahraman et al. Citation2013). Vicia are represented by 66 species, 27 subspecies and 29 varieties in Turkey (Başbağ et al. Citation2013; Binzat et al. Citation2014).
The genus Vicia is widely used in the world and in our country. Many taxa are used in the food and feed industry because they are very rich in terms of nutritional value. The taxa are also used as feed plants in the agricultural area, grassland plants, green manure plants and used in the cultivation season in the fallow soil. In addition, some taxa are also grown as ornamental plants in parks and gardens. There are also taxa used as bioindicators to remove oil pollution, especially in the seas (Kahlaoui et al. Citation2009; Gedik et al. Citation2013; Başbağ et al. Citation2013; Büyükkartal et al. Citation2013; Kahraman et al. Citation2013; Binzat et al. Citation2014).
Different diploid chromosome number counts have been reported in the genus Vicia. In the genus, the most frequent chromosome numbers are 2n = 2x = 12 and 14. The number of somatic chromosomes of the taxa belonging to the genus Vicia varies from 2n = 6 to 2n = 42, e.g. 2n = 6, 10, 12, 14, 18, 22, 24, 26, 28 and 42 (Yefimov Citation1988; Şahin and Babaç Citation1990; Şahin et al. Citation1996; Meriç and Dane Citation1999; Bağcı and Şahin Citation2000; Rahiminejad et al. Citation2000; Tabur et al. Citation2001; Karadağ and Büyükburç Citation2003; Sevimay et al. Citation2005; Çeliktaş et al. Citation2006; Gianfranco et al. Citation2008; Namazi et al. Citation2008; Kahlaoui et al. Citation2009; Tabur et al. Citation2009; Arslan et al. Citation2012; Gedik et al. Citation2013).
Karyotype asymmetry is widely used in comparative cytotaxonomy. The symmetrical karyotypes are mainly derived from median and submedian chromosomes with approximately the same size, whereas the asymmetric karyotypes are mainly derived from subtelocentric and telocentric chromosomes. The asymmetry degrees may also increase depending on differences in relative sizes. In parallel, the karyotype heterogeneity also increases. Karyotype asymmetry consists of two parts as intrachromosomal asymmetry and interchromosomal asymmetry (Peruzzi and Eroglu Citation2013). The parameters of MCA (Peruzzi and Eroglu Citation2013) and CVCL (Paszko Citation2006) are perfectly suited to measure intrachromosomal asymmetry and interchromosomal asymmetry, respectively.
In the present study, three sections of the genus Vicia were investigated karyologically to determine the detailed chromosome measurements and to compare with earlier results. New information on the karyotype of the examined species growing in Turkey has been obtained by karyotype analysis, which might help a better understanding of their taxonomic treatment.
Materials and methods
Seeds of seven Vicia taxa were collected from central Anatolia in Turkey. These taxa are V. articulata, V. cassubica, V. villosa subsp. villosa in sect. Cracca; V. noeana var. noeana, V. sativa subsp. sativa, V. peregrina in sect. Vicia and V. caesarea in sect. Ervum. The localities of the taxa used in the study are given in .
Table 1. Localities and collector numbers of studied Vicia.
The seeds were germinated between wet filter papers in Petri dishes. The root tips were pretreated in α-bromonaphthalene for 16 h at 4°C, then fixed in absolute alcohol-acetic acid (3:1) for 24 h, and then kept in 70% ethanol at 4°C in the refrigerator. The root tips were hydrolyzed in 1 N HCl for 12 min at room temperature, then stained in aceto-orcein for 2 h at room temperature, and finally squashed in 45% acetic acid. The permanent slides were made by mounting in Depex (Esra et al. Citation2008, Martin et al. Citation2011, Citation2015). The observations of the mitotic plates were made using an Olympus BX-53 microscope (Olympus Corporation, Tokyo, Japan) and photographed with an Olympus DP72 digital camera.
Chromosome properties including long arm length (L), short arm length (S), total chromosome length (L + S), arm ratio (L/S), relative length (RL) and centromeric index (CI = S/[L + S] × 100) were measured using Software Image Analyses (Bs200ProP). The determination of centromeric position as median, submedian and/or subtelocentric followed Levan et al. (Citation1964). The karyotype asymmetries were classified as the Stebbins classification (a quali-quantitative method), intrachromosomal asymmetry using MCA, AsK, TF, Syi, A1 and A and interchromosomal asymmetry using CVCL, Rec and A2 (for details see Paszko Citation2006; Eroğlu et al. Citation2013; Peruzzi and Eroglu Citation2013). Finally, asymmetry parameters were compared with Pearson correlation.
Results
The chromosomes of the genus Vicia are large enough and the centromere regions are quite evident. For this reason, chromosome types can be easily distinguished. The characteristics of mitotic metaphase chromosomes are given below according to the measurement data.
Sect. Cracca
Vicia articulata
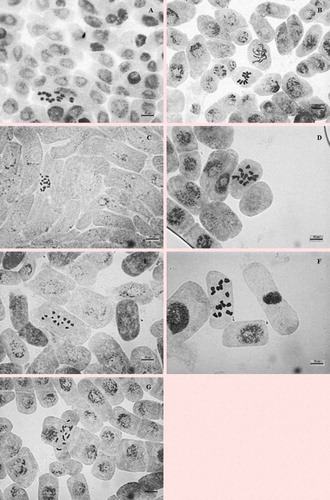
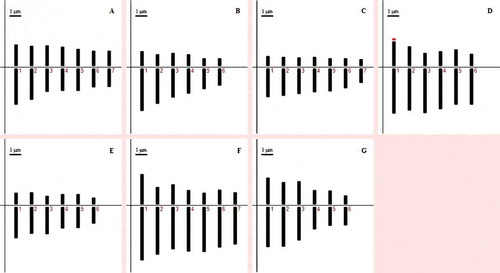
The chromosome number of V. articulata is 2n = 14 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 2.93, 4.99, 3.74 and 26.18 μm, respectively. The chromosome arm rates, centromeric index and relative length are 1.20–1.69, 37.27–46.84 and 11.18–19.08, respectively. The karyotype formula is 2n = 14 = 7m (). ) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 1A, 13.28 and 20.20, respectively.
Table 2. The measurement data of chromosome pairs of species of section Cracca.
Vicia cassubica
The chromosome number of V. cassubica is 2n = 12 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 2.09, 4.73, 3.28 and 19.69 μm, respectively. The chromosome arm rates, centromeric index and relative length are 2.12–2.99, 25.06–32.06 and 10.60–24.03, respectively. The karyotype formula is 2n = 12 = 6sm (). ) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 4B, 42.15 and 30.00, respectively.
Vicia villosa subsp. villosa
The chromosome number of V. villosa subsp. villosa is 2n = 14 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 1.86, 3.36, 2.75 and 19.25 μm, respectively. The chromosome arm rates, centromeric index and relative length are 2.08–2.96, 25.32–32.43 and 9.64–17.47, respectively. The karyotype formula is 2n = 14 = 7sm (). )) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 4A, 45.51 and 18.55, respectively.
Sect. Vicia
Vicia noeana var. noeana
The chromosome number of V. noeana var. noeana is 2n = 12 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 4.23, 6.05, 5.01 and 30.08 μm, respectively. The chromosome arm rates, centromeric index and relative length are 1.80–3.33, 23.06–35.70 and 14.07–20.12, respectively. The karyotype formula is 2n = 12 = 5sm + 1st (). The satellite is observed in the first chromosome pair. ) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 3A, 41.68 and 12.58, respectively.
Table 3. The measurement data of chromosome pairs of species of section Vicia.
Vicia sativa subsp. sativa
The chromosome number of V. sativa subsp. sativa is 2n = 12 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 2.07, 3.72, 2.96 and 17.75 μm, respectively. The chromosome arm rates, centromeric index and relative length are 1.98–3.17, 24.04–33.70 and 11.65–20.99, respectively. The karyotype formula is 2n = 12 = 5sm + 1st (). ) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 3A, 40.52 and 19.56, respectively.
Vicia peregrina
The chromosome number of V. peregrina is 2n = 14 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 4.32, 7.21, 5.34 and 37.38 μm, respectively. The chromosome arm rates, centromeric index and relative length are 1.75–3.73, 21.18–36.34 and 11.56–19.29, respectively. The karyotype formula is 2n = 14 = 5sm + 2st (). ) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 3A, 44.62 and 17.42, respectively.
Sect. Ervum
Vicia caesarea
The chromosome number of V. caesarea is 2n = 12 ()). The smallest chromosome length, the largest chromosome length, the mean chromosome length and the total haploid length are 2.39, 5.78, 4.08 and 24.46 μm, respectively. The chromosome arm rates, centromeric index and relative length are 1.32–2.02, 33.05–43.01 and 9.78–23.64, respectively. The karyotype formula is 2n = 12 = 4m + 2sm (). ) shows the monoploid ideogram. The values of the Stebbins classification, intrachromosomal asymmetry (MCA) and interchromosomal asymmetry (CVCL) are 2B, 22.45 and 35.18, respectively.
Table 4. The measurement data of chromosome pairs of species of section Ervum.
Discussion
The detailed chromosome measurements and karyotype asymmetry are significant data for cytotaxonomy of plant. Wild and cultivated species are used because of their economic importance in family Fabaceae (Jha et al. Citation2017). Thus it is possible to make phylogenetic comparisons of ancient and new crops. Recently, synthetic polyploids are also used in chromosomal data of genus Vicia (Pavlíková et al. Citation2017). Phylogenetic and cytogenetic data may be useful for conservation of genetic diversity and future crop breeding methods.
In –, the chromosome numbers of V. cassubica, V. noeana var. noeana, V. sativa subsp. sativa and V. caesarea are shown as 2n = 12. It was reported that the chromosome numbers were 2n = 10 in V. sativa L. subsp. incisa (Bieb.) Arc.; 2n = 12 in V. cassubica, V. noeana, V. noeana var. noeana, V. noeana Boiss. & Reut. ex Boiss. var. megalodonta Rech. Fil., V. sativa, V. sativa subsp. sativa, Vicia sativa L. subsp. nigra (L.) Ehrh. and V. caesarea; 2n = 14 in V. noeana var. noeana, V. sativa subsp. nigra and V. sativa subsp. incisa (Yefimov Citation1988; Şahin and Babaç Citation1990; Şahin et al. Citation1996; Meriç and Dane Citation1999; Bağcı and Şahin Citation2000; Rahiminejad et al. Citation2000; Sevimay et al. Citation2005; Gianfranco et al. Citation2008; Namazi et al. Citation2008; Kahlaoui et al. Citation2009; Arslan et al. Citation2012; Gedik et al. Citation2013). In Çeliktaş et al. (Citation2006), the chromosome number was 2n = 10 in wild V. sativa and 2n = 12 in cultured V. sativa. All reports include both similar and dissimilar results with our results.
In and , the chromosome numbers of V. articulata, V. villosa subsp. villosa and V. peregrina are shown as 2n = 14. It was reported that the chromosome numbers are 2n = 12 in V. peregrina; 2n = 14 in V. articulata, V. villosa, V. villosa Roth subsp. dasycarpa (Ten.) Stank. and V. peregrina (Rahiminejad et al. Citation2000; Tabur et al. Citation2001, Citation2009; Namazi et al. Citation2008; Kahlaoui et al. Citation2009). The reports include both similar and dissimilar results to our results. Also, Pavlíková et al. (Citation2017) showed the 26 diploid (2n = 2x = 14), 25 natural tetraploid (2n = 4x = 28) and 27 synthetic tetraploid (2n = 4x = 28) in basic species V. cracca of sect. Cracca. They reported that the comparison of synthetic and natural polyploids provide understanding of the results of polyploidization and separate the effects of the duplication from evolution. Polyploidy originates by autopolyploidy (genome duplication of one species) and allopolyploidy (genome duplication with different species) and has played a major role in the speciation and evolution of higher plants (Demirci Kayıran and Özhatay Citation2017). The polyploidy rates increase with factors such as climate shifts, glaciations, high latitudes and high altitudes (Metzgar et al. Citation2016) In future studies, it is necessary to compare diploids and synthetic polyploids for all Vicia species.
The chromosome morphologies belonging to the studied taxa are different. Turkey is a very convenient area for genetic variations because of its location in different plant geographical regions which are Mediterranean, Euro-Siberian and Irano-Turanian. The genetic variations in Vicia in Turkey, which are clearly detectable, are in the chromosome morphologies and chromosome numbers. The haploid karyotype formulae are 7m in V. articulata, 6sm in V. cassubica, 7sm in V. villosa subsp. villosa, 5sm + 1st in V. noeana var. noeana, 5sm + 1st in V. sativa subsp. sativa, 5sm + 2st in V. peregrina and 4m + 2sm in V. caesarea. As an interesting note, sect. Cracca and Ervum only have median and submedian chromosomes, with no subtelocentric ones, and sect. Vicia only has submedian and subtelocentric chromosomes, with no median ones. A satellite was only observed in the first pair of chromosomes of V. noeana var. noeana. In literature, different chromosome formulae have been reported.
The karyotype formula of V. articulata was 1m + 2sm + 4st with satellite in the first and fourth chromosome pairs (Tabur et al. Citation2001). The karyotype formula of V. villosa subsp. villosa was 1m + 6sm with satellite in the seventh chromosome pair (Namazi et al. Citation2008). The karyotype formula of V. villosa subsp. darycarpa was 7sm with satellite in the sixth chromosome pair (Tabur et al. Citation2001). The karyotype formulae of V. noeana var. noeana were 6m (Karadağ and Büyükburç Citation2003), 3sm + 3st, 2sm + 4st, 6sm with satellite in the first chromosome pair (Gianfranco et al. Citation2008) and 1m + 3sm + 2st (Gedik et al. Citation2013). The satellite in the first chromosome pair is compatible with this study, but not the karyotype formula. The karyotype formula of V. noeana var. megalodonta was 5sm + 1st with satellites in the third and sixth chromosome pairs (Gedik et al. Citation2013). The karyotype formula is compatible with this study, but not the satellite. The karyotype formulae of V. sativa subsp. sativa were 1m + 5sa with satellites in the second and third chromosome pairs (Namazi et al. Citation2008) and 2sm + 3st + 1t (Gedik et al. Citation2013). The karyotype formulae of V. sativa subsp. nigra and V. sativa subsp. incisa were 1m + 1sm + 4a (Arslan et al. Citation2012) and 1sm + 6st (Meriç and Dane Citation1999), respectively. The karyotype formulae of V. peregrina were 1m + 3st + 3t and 1m + 4st + 2t with satellites in the first and third chromosome pairs (Gianfranco et al. Citation2008) and 7st with satellite in the third chromosome pair (Tabur et al. Citation2009). The karyotype formula of V. caesarea was 1m + 5sm (Şahin et al. Citation1996). Our results are different from all reports given above. It was reported that the differences in chromosome morphologies may contribute to the variation of the genera, sections and species (Altay et al. Citation2017).
The intrachromosomal asymmetry gradually increases with centromere shift from median point to terminal point. The values of AsK, TF, Syi, A1, A, and MCA are parameters used in the calculation of intrachromosomal asymmetry. The interchromosomal asymmetry increases with more heterogeneity of chromosome sizes. Rec, A2 and CVCL are parameters used in the calculation of intrachromosomal asymmetry (Peruzzi and Eroglu Citation2013).
In sect. Cracca, V. articulata is the most symmetrical karyotype, while V. villosa subsp. villosa is the most asymmetrical karyotype according to the MCA, AsK, TF, Syi, A1, and A. Whereas V. villosa subsp. villosa is the most symmetrical karyotype, V. cassubica is the most asymmetrical karyotype in CVCL, Rec, and A2 ().
Table 5. The asymmetry index values of Vicia taxa.
In sect. Vicia, V. sativa subsp. sativa is the most symmetrical karyotype, while V. peregrina is the most asymmetrical karyotype according to the MCA, AsK, TF, Syi, A1, and A. Whereas V. noeana var. noeana is the most symmetrical karyotype, V. sativa subsp. sativa is the most asymmetrical karyotype in CVCL together with only A2. As a small difference from the other parameters, V. noeana var. noeana is the most symmetrical karyotype, while V. peregrina is the most asymmetrical karyotype in Rec value ().
In all sections, V. articulata is the most symmetrical karyotype, while V. villosa subsp. villosa is the most asymmetrical karyotype according to the MCA, AsK, TF, Syi, A1, and A. However, the asymmetrical karyotypes are different in interchromosomal asymmetries. While V. noeana var. noeana is the most symmetrical karyotype in CVCL, Rec, and A2. V. caesarea is the most asymmetrical karyotype in only CVCL and A2. Unlike all parameters, V. cassubica is the most asymmetrical karyotype in Rec value ().
Intrachromosomal asymmetry follows the same trends as the taxa showing symmetrical and asymmetric karyotypes. In , the MCA value showed perfect positive and negative correlations with AsK (r = 0.998), TF (r = − 0.998), Syi (r = − 0.998), A1 (r = 0.999) and A (r = 0.998) for intrachromosomal asymmetry. In interchromosomal asymmetry, it is the same as the taxa showing symmetrical and asymmetric karyotypes. In , while the CVCL value showed the perfect positive correlation with A2 (r = 0.999), it showed weaker correlation with Rec (r = − 0.826).
Table 6. Pearson correlation for asymmetry indices.
The very weak negative correlations were determined in an interchromosomal asymmetry against an intrachromosomal asymmetry as MCA–CVCL (r = − 0.380), Syi–Rec (r = − 0.363) and A1–A2 (r = − 0.333) and ( and Figure 3). It was likely that the weak negative correlations are due to parameters of interchromosomal asymmetry. It was reported that a parameter of interchromosomal asymmetry gives reliable results with a parameter of intrachromosomal asymmetry (Peruzzi and Eroglu Citation2013). gives scatter diagrams of MCA–CVCL and Syi–Rec. The diagrams are like mirror images of each other. This is because the MCA–CVCL values increase with increasing asymmetry; conversely, Syi–Rec values decrease with increasing asymmetry (Eroğlu et al. Citation2013; Altay et al. Citation2017).
Figure 3. Scatter diagram between MCA–CVCL and Syi–Rec: Sect. of Cracca, Vicia and Ervum. (a) V. articulata; (b) V. cassubica; (c) V. villosa subsp. villosa; (d) V. noeana var. noeana; (e) V. sativa subsp. sativa; (f) V. peregrina; (g) V. caesarea.
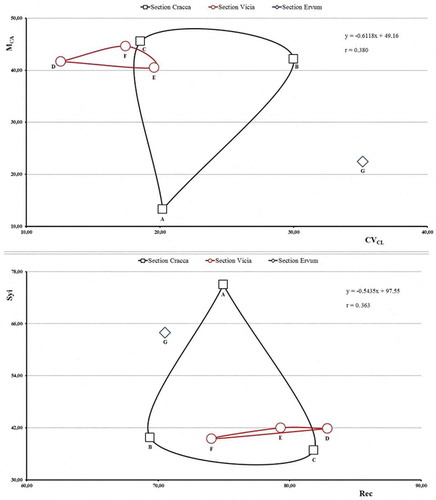
In , the sections of genus Vicia have different degrees of karyotype asymmetry: sect. Cracca with a balance between interchromosomal and intrachromosomal, sect. Vicia with higher intrachromosomal, sect. Ervum with higher interchromosomal. On the other hand, more chromosomal data will give more accurate results.
In this study, detailed chromosome measurements and karyotype asymmetry degrees are determined for V. articulata, V. cassubica, V. villosa subsp. villosa, V. noeana var. noeana, V. sativa subsp. sativa, V. peregrina and V. caesarea. The diploid chromosome numbers and basic chromosome numbers are 2n = 12, 14 and x = 6, 7. In many genera and genus Vicia, the basic chromosome numbers are x = 5, 6, 7 and the most common diploid numbers are 2n = 12, 14 (Gianfranco et al. Citation2008; Gedik et al. Citation2013; Tanaomi et al. Citation2016). All taxa have relatively large chromosomes, between 1.86 and 7.21 μm. The little variations were showed in size of the chromosomes. The karyotypes of V. articulata are quite symmetric, because it has completely median chromosomes. In all results, chromosomes vary from median to subterminal and karyotypes vary from symmetric to asymmetric. The variation of interspecific, intersection and intergeneric may increase with diversity in karyotype asymmetry degrees and karyotype formulae.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Altay D, Eroğlu HE, Hamzaoğlu E, Koç M. 2017. Karyotype analysis of some taxa of Dianthus section Verruculosi (Caryophyllaceae, Sileneae). Turk J Bot. 41:367–374.
- Arslan E, Ertuğrul K, Öztürk AB. 2012. Karyological studies of some species of the genus Vicia L. (Leguminosae) in Turkey. Caryologia. 65:106–113.
- Bağcı E, Şahin A. 2000. A numerical cytotaxonomic study on some Vicia L. taxa. Herb J Syst Bot. 7:143–160.
- Başbağ M, Hoşgören H, Aydın A. 2013. Vicia taxa in the flora of Turkey. Anadolu Tarım Bilimleri Dergisi. 28:59–66.
- Binzat OK, Kahraman A, Doğan M. 2014. Pollen morphology of some taxa of Vicia L. subgenus Vicilla (Schur) Rouy (Fabaceae) from Turkey. Plant Syst Evol. 300:1867–1876.
- Büyükkartal HN, Çölgeçen H, Pınar NM, Erdoğan N. 2013. Seed coat ultrastructure of hard-seeded and soft-seeded varieties of Vicia sativa. Turk J Bot. 37:270–275.
- Çceliktaş N, Can E, Hatipoğlu R, Avci S. 2006. Comparison between a wild population and cultivar of common vetch (Vicia sativa L., Fabaceae) on cytological and agronomic characteristics. New Zeal J Agr Res. 49:389–393.
- Demirci Kayıran S, Özhatay FN. 2017. A karyomorphological study on the genus Muscari Mill. growing in Kahramanmaraş (Turkey). Turk J Bot. 41:289–298.
- Eroğlu HE, Şimşek N, Koç M, Hamzaoğlu E. 2013. Karyotype analysis of some Minuartia L. (Caryophyllaceae) taxa. Plant Syst Evol. 299:67–73.
- Esra M, Akan H, Ekici M, Aytaç Z. 2008. Karyomorphological studies on section Bucerates Boiss. of Trigonella L. (Leguminosae) from Turkey. Caryologia. 61:225–236.
- Gedik O, Kıran Y, Şahin A. 2013. Vicia L. cinsine ait bazı taksonların karyolojik yönden araştırılması. Bitlis Eren Üniversitesi Fen Bilimleri Dergisi. 2:12–20.
- Gianfranco V, Ravalli C, Cremonini R. 2008. The karyotype as a tool to identify plant species: Vicia species belonging to Vicia subgenus. Caryologia. 61:300–319.
- Jaaska V. 2005. Isozyme variation and phylogenetic relationships in Vicia subgenus Cracca (Fabaceae). Ann Bot. 96:1085–1096.
- Jha TB, Saha PS, Adak M, Jha S, Roy P. 2017. Chromosome morphometric analysis of Indian cultivars of Lens culinaris Medik. using EMA based Giemsa staining method. Caryologia. 70:270–283.
- Kahlaoui S, Walker DJ, Correal E, Martínez-Gómez P, Hassen H, Bouzid S. 2009. The morphology, chromosome number and nuclear DNA content of Tunisian populations of three Vicia species. Afr J Biotechnol. 8:3184–3191.
- Kahraman A, Binzat OK, Doğan M. 2013. Plant pollen morphology of some taxa of Vicia L. subgenus Vicia (Fabaceae) from Turkey. Plant Syst Evol. 299:1749–1760.
- Karadağ Y, Büyükburç U. 2003. Karyotype analysis of some legume species (Vicia noeana Boiss. and Lathyrus sativus L.) collected from native vegetation. Pak J Biol Sci. 6:377–381.
- Kupicha FK. 1976. The infrageneric structure of Vicia L. Notes Roy Bot Gard Edinburgh. 34:287–326.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52:201–220.
- Lewis GM, Schrire B, Mackinder B, Lock M. 2005. Legumes of the world. London: Kew Press.
- Martin E, Akan H, Ekici M, Aytac Z. 2011. Karyotype analysis of ten sections of Trigonella (Fabaceae). Comp Cytogenet. 5:105–121.
- Martin E, Doğan B, Duran A, Coşkun F, Altınordu F. 2015. Contribution to the cytotaxonomic knowledge of 10 species of Klasea Cass. (Asteraceae) fromTurkey. Caryologia. 68:330–338.
- Maxted N. 1993. A phenetic investigation of Vicia L. subgenus Vicia (Leguminosae–vicieae). Bot J Linn Soc. 111:155–182.
- Maxted N, Hawkes JG. 1997. Selection target taxa. In: Maxted N, Ford-Lloyd BV, Hawkes JG, editors. Plant genetic conservation: the in situ approach. London: Chapman and Hall; p. 43–68.
- Meriç Ç, Dane F. 1999. Karyological studies on Vicia sativa ssp. incisa (Bieb.) Arc. var. incisa. Turk J Bot. 23:63–67.
- Metzgar J, Stamey M, Ickert-Bond S. 2016. Genetic differentiation and polyploid formation within the Cryptogramma crispa complex (Polypodiales: Pteridaceae). Turk J Bot. 40:231–240.
- Miller JT, Murphy DJ, Brown GK, Richardson DM, González-Orozco CE. 2011. The evolution and phylogenetic placement of invasive Australian Acacia species. Divers Distrib. 17:848–860.
- Namazi LG, Badrzadeh M, Asghari-Zakaria R. 2008. Karyotype of several Vicia species from Iran. Asian J Plant Sci. 7:417–420.
- Paszko B. 2006. A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 258:39–48.
- Pavlíková Z, Paštová L, Münzbergová Z. 2017. Synthetic polyploids in Vicia cracca: methodology, effects on plant performance and aneuploidy. Plant Syst Evol. 303:827–839.
- Peruzzi L, Eroglu HE. 2013. Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet. 7:1–9.
- Rahiminejad MR, Ehtemam MH, Neishaboori A. 2000. Cytotaxonomic studies of some Iranian Vicia species (Fabacaea). J Sci I R Iran. 11:1–5.
- Şahin A, Babaç MT. 1990. Cytotaxonomic studies of some Vicia L. species in eastern and south-eastern Anatolia I. Turk J Bot. 14:124–138.
- Şahin A, Çobanoğlu D, Gür N. 1996. The morphological, karyological and palynological features of Vicia caeserea Boiss. & Bal. (Fabaceae). Turk J Bot. 201:31–36.
- Sevimay CS, Güloğlu D, Khawar KM. 2005. Karyotype analysis of eight Turkish vetch (Vicia sativa L.) cultivars. Pakistan J Bot. 37:313–317.
- Tabur S, Cesur A, Özkul H. 2009. Karyology of seven Fabaceae taxa from Turkey. J Appl Biol Sci. 3:49–53.
- Tabur S, Civelek S, Bagci E. 2001. Cytotaxonomic studies on some Vicia L. species growing in eastern Mediterranean and southern Aegean regions, I. Acta Bot Gallica. 148:159–174.
- Tanaomi N, Jonoubi P, Rad AC, Majd A, Ranjbar M. 2016. Embryology of Onobrychis persica Sirj. and Rech.f. (Fabaceae) and its systematic implications. Caryologia. 69:256–266.
- Yefimov KF. 1988. Caryological study of the species of the genus Vicia (Fabaceae) from the central Caucasus. Bot Žurnal (Moscow & Leningrad). 73:641–651.