Abstract
Current diagnostic techniques for the detection of the small hive beetle (SHB), Aethina tumida are limitedly available and not cost effective. More sensitive pragmatic methods are preferred for early detection. To improve diagnostics, we focused on sampling techniques for SHB frass, as an indicator for SHB presence in a honey bee colony. In this study, we successfully tested a novel approach of employing swab sample collection of frass for real-time PCR detection of SHB.
Small hive beetle (SHB), Aethina tumida Murray, is an invasive honey bee pest that has become established worldwide, with the potential to spread even further (Neumann et al., Citation2016; Cornelissen et al., Citation2019). Honey bee colonies are attractive to free flying, SHB individuals. All known invasions have first been detected in honey bee hives. Commonly, visual screening of honey bee colonies is used to detect SHBs (Cornelissen & Neumann, Citation2018). This is however time consuming and has a low success rate. PCR detection seems a better option, but both field collection & laboratory methods are yet to be optimized (Schäfer et al., Citation2019). One such optimisation could be the detection of SHB frass, present in the bee hive. Here, we define frass as solid faecal excrement deposited by SHBs. Frass could serve like a fingerprint, showing the past or current presence of SHB in the hive. We hypothesize that SHB DNA can be detected using swabs to collect frass samples and with the potential to increase the options for early detection of SHB in hives. Furthermore, we suggest improvements for current DNA extraction and real time PCR method for the detection of SHB.
We obtained frass samples from the bee lab at Auburn University, College of Agriculture. SHBs (n = 2) were put in a closed petri dish (Ø 100 mm) with Whatman cellulose filter paper (Merck, Darmstadt, Germany) on the bottom. After 24 hrs, the SHBs were removed, the filter papers were stored at room temperature and sent over to our laboratory (Wageningen Plant Research, Wageningen, the Netherlands). From one filter paper, various frass samples (n = 3) were punched out (Ø 4 mm) using a sample puncher (Vanem, The Netherlands). Also, single frass samples (n = 2) were collected using a flocked swab (FLOQSwab, Copa961C, Copan, Brescia, Italy). Visual confirmation of SHB frass sample collection was performed by checking the samples under a stereo-microscope (magnification 40x). More samples were collected at the Istituto Zooprofilattico Del Mezzogiorno (Calabria, Italy). Six SHBs were held in plastic storage containers and fed sugar water (40% weight ratio) ad libitum. After three days, all visual frass (n = 11), were secured using one flocked swab per frass. Additionally, the gut content from a squashed adult beetle was collected using a swab (n = 2). Also, the exoskeleton of SHBs (n = 5) was sampled using a swab. To mimic biological samples, swab samples (n = 7) were taken on the inside of an empty used hive part from bees@wur at Wageningen, the Netherlands (lid, brood box, top bars, and bottom board). Furthermore, we swab-sampled two possible inhibitors; Bee repellent (Onetti fabi-spray, Italy) (n = 2) and honey (n = 1). Simple DNA extraction was performed using a Nexttec Kit for Tissue & Cells (nexttec Biotechnologie GmbH, Germany) for swabs and filter paper punches. The swabs were cut to fit in a 1.5 ml eppendorf tube. Together with 140 µl lysis buffer G, 10 µl Prot K and 1.5 µl DTT, the punches or swabs were incubated for 3 hrs at 56 °C at 1000 rpm. After the lysis step, the swab tips were turned upside down and a short spin released the buffer. The kit was then used according to the manufacturer’s instructions. Regarding molecular detection of SHB, we used an SHB-specific real-time PCR assay described by Li et al. (Citation2018). We modified the probe with Locked Nucleic Acid (LNA)-bases, to obtain a more sensitive and cost-effective assay. Base notation for LNA bases is shown as + N, Atum-LNA_P: 5′-[6FAM]-TA + TTTGCTAT + TATA + GCCGGATTT + GT-[IABkFQ]-3′. As a positive control, a synthetic double stranded verified DNA fragment (gBlock) was designed, containing the corresponding COI gene sequence with primers and probe binding sites. The gBlock-COI-Li (5′-CATCTATTGATATTATTCTACA TGATACTTACTACGTAGTAGCCCATTTCCATTATG TATTATCTATAGGAGCAGTATTTGCTATTATAGCCG GATTTGTTCAATGATTCCCATTAATTACAGGATTAA CTTTAAATAGAAATTATT-3′) was 147 bp long. The primers, probe and gBlock were synthesized by Integrated DNA Technologies (Coralville, IA, USA). A standard curve was made by serial 10-fold dilution of the gBlock-COI-Li in TE buffer from 106 to 1 copy per 1 µl. Inhibition of background material was investigated by spiking 100 copies gBlock-COI-Li into non target swab samples from the hive, bee repellent and honey. The real-time PCR assay was performed in a 25 µl volume consisting of 12.5 µl PerfeCta qPCR ToughMix Low ROX (Quantabio, Beverly MA, USA), primers (300 nM) and probe (100 nM), 5 µl of DNA or 1 µl gBlock, and PCR water. Thermal cycling conditions consisted of 2 min 95 °C, 40 cycles of 95 °C for 15 s and 60 °C for 1 min. No-Template-Control (NTC) reactions were included. The Quantstudio 12k Flex was used and thresholds were set automatically by the software v1.3. The technical sensitivity of the real-time PCR was evaluated using the calculated standard curve, which gave a linear curve for 106 to 10 copies gBlock-COI-Li with correlation coefficient R2 = 0.994 and efficiency of amplification E = 101.17. The slope was −3.294 and Y-Intercept 37.989 (see supplement ). We compared Ct values for the different sample types using a univariate General Linear Model (GLM) with Ct value as a dependent factor and sample types as a fixed factor. Estimated Marginal Means were calculated with Sidak as a confidence interval adjustment.
Figure 1. Real-time PCR results of five different sample types from small hive beetle: exoskeleton swabs (n = 5), gut content swabs (n = 2), paper punch with frass (n = 3), frass swabs from paper (n = 2) frass swabs from container (n = 11). The Ct value represents the Ct mean of each sample type and the top bar indicates the standard error. Lower case letters indicate significant differences (P < 0.05) between sampling types.
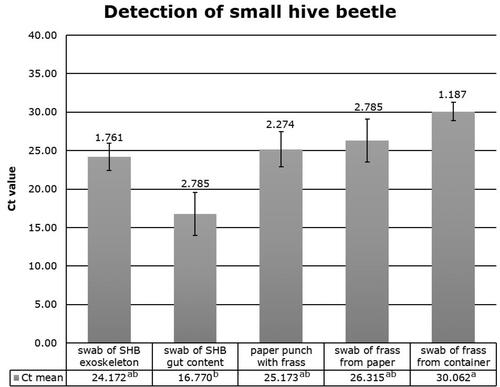
While all NTC were negative, the diagnostic method was able to accurately identify SHB in all SHB derived sample types (). Constituents of bee hive dirt, bee repellent, or honey did not inhibit the diagnostic assay since the expected value of Ct = 32.00 was not affected (supplement ). The model result was significant (df = 4, F5.814, P = 0.003). A pairwise comparison revealed gut sample Ct-values were significantly lower (P < 0.01) than swab samples taken from the container. Ct values of other treatments were not significantly different from one another in a pairwise comparison (P > 0.05).
SHBs are attracted to honey bee colonies, the principle by which sentinel colonies are used for early detection in uninvaded regions (Schäfer et al., Citation2019). And where SHBs reside in colonies they are likely to leave traces of their presence (Cornelissen, unpublished data). Our study shows that SHB frass is suitable for molecular diagnostics. Small amounts of targets in SHB frass of different origins and composition could be detected with a modified real-time PCR assay. Our modification (LNA) shows potential as it offers an accurate, specific and cost-effective alternative, while other assays currently used are possibly outdated (Li et al., Citation2018). Furthermore, by using flocked swabs in a simple non-destructive sampling method, combined with a nearly loss-free DNA preparation of small amounts of starting material, we’ve expanded the potential for SHB diagnostics and bee diseases in general.
Figure S1
Download MS Word (172.6 KB)Acknowledgements
We thank the following persons for discussion and assistance and for providing beetles and frass: Madeleine van Eijk, Giovanni Formato, Giovanni Frederico, Marc Hendriks, Peter Neumann and Geoff Williams.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
is available via the ‘Supplementary’ tab on the article’s online page (http://dx.doi.org/10.1080/00218839.2021.1914964).
Additional information
Funding
References
- Cornelissen, B., & Neumann, P. (2018). How to catch a small beetle: Top tips for visually screening honey bee colonies for small hive beetles. Bee World, 95(3), 99–102. https://doi.org/https://doi.org/10.1080/0005772X.2018.1465374
- Cornelissen, B., Neumann, P., & Schweiger, O. (2019). Global warming promotes biological invasion of a honey bee pest. Global Change Biology, 25(11), 3642–3655. https://doi.org/https://doi.org/10.1111/gcb.14791
- Li, D., Waite, D. W., Fan, Q. H., George, S., Semeraro, L., & Blacket, M. J. (2018). Molecular detection of small hive beetle Aethina tumida Murray (Coleoptera: Nitidulidae): DNA barcoding and development of a real-time PCR assay. Scientific Reports, 8(1), 1–13. https://doi.org/https://doi.org/10.1038/s41598-018-27603-x
- Neumann, P., Pettis, J. S., & Schäfer, M. O. (2016). Quo vadis Aethina tumida? Biology and control of small hive beetles. Apidologie, 47(3), 427–466. https://doi.org/https://doi.org/10.1007/s13592-016-0426-x
- Schäfer, M. O., Cardaio, I., Cilia, G., Cornelissen, B., Crailsheim, K., Formato, G., Lawrence, A. K., Le Conte, Y., Mutinelli, F., Nanetti, A., Rivera-Gomis, J., Teepe, A., & Neumann, P. (2019). How to slow the global spread of small hive beetles. Aethina tumida. Biological Invasions, 21(5), 1451–1459. https://doi.org/https://doi.org/10.1007/s10530-019-01917-x
