ABSTRACT
Nakamigawaia is a poorly understood genus of Aglajidae sea slugs with only two species formally ascribed. In this paper we explore new morpho-anatomical characters using stereo and scanning electron microscopy and employ different molecular approaches (a cytochrome c oxidase sub-unit I gene phylogeny, the Automatic Barcode Gap Discovery species delimitation method, and genetic distances) to compare specimens across the geographical span of the genus and from two distinct chromatic morphotypes occurring in the Western Pacific (blackish morph and white-dotted morph). Our results support the conspecificity of these two morphs and show they belong to an undescribed species here named Nakamigawaia nakanoae sp. nov. The species differs from the type species of the genus, N. spiralis, by the presence of a distinct open-dilated shell and differs from its Western Atlantic congener N. felis by subtle differences in the shell, male reproductive system and caudal lobes. Genetically (COI uncorrected p-distance) the two species (N. nakanoae and N. felis) are 18.8–20.1% distinct. The definition of the genus Nakamigawaia is discussed and the current assignment to the latter of lineages other than the type species is questioned.
Introduction
The family Aglajidae is the second most diverse of the gastropod Heterobranchia order Cephalaspidea, with an estimated 85 valid species worldwide divided into 15 genera (Zamora-Silva and Malaquias Citation2018). These marine slugs inhabit tropical and temperate waters predominantly at shallow depths where they are found crawling on sandy bottoms, coral rubble and algae (Burn and Thompson Citation1998; Gosliner et al. Citation2008; Camacho-García et al. Citation2014; Zamora-Silva and Malaquias Citation2018), yet at least three species have been reported from deeper bathymetries including abyssal depths (Zamora-Silva and Malaquias Citation2018; Chaban et al. Citation2019). Aglajids are predators feeding mostly upon vagile prey such as other sea slugs, nematodes, polychaetes and small fish (Zamora-Silva and Malaquias Citation2016).
One of the genera of Aglajidae is the enigmatic Nakamigawaia Kuroda and Habe, Citation1961. The genus was introduced for the species N. spiralis Kuroda and Habe, Citation1961 from Japan (type locality: Kasajima beach, Sagami Bay) and originally included in the family Chelidonuridae Kuroda and Habe Citation1961 (presently regarded as a synonym of Aglajidae; MolluscaBase Citation2021). Kuroda and Habe (Citation1961) provided only a brief description, in Japanese, of the colouration of the animal and an illustration of the shell. The definition of the genus and species remained largely elusive, and, for example, Rudman (Citation1978) and Gosliner (Citation1980) did not consider this genus in their dedicated accounts on the systematics of the aglajid slugs.
Baba (Citation1985) was the first author to provide a detailed description of the type species N. spiralis, recognising at the same time similarities to representatives of the Aglajidae genus Melanochlamys, which led the author to firmly suggest the inclusion of the genus Nakamigawaia in the family. The validity of Nakamigawaia and its inclusion in the Aglajidae have been corroborated by Camacho-García et al. (Citation2014) and Zamora-Silva and Malaquias (Citation2018) based on molecular phylogenetics. In addition, Camacho-García et al. (Citation2014) first suggested a close relationship between N. spiralis (based on specimens from Papua New Guinea and the Philippines) and the morphologically similar tropical western Atlantic species ‘Aglaja’ felis described by Marcus and Marcus (Citation1970) from Puerto Rico, which led the authors to reassign the latter species to the genus Nakamigawaia.
Despite the putative phylogenetic evidence for a common generic affiliation of the Atlantic and Pacific species, the occurrence of substantial differences between their shells (convex, dilated with an open whorl in ‘A’. felis, and spiralled in N. spiralis), led Ortea et al. (Citation2014) to argue these species warranted a different generic assignment, and introduced the genus name Migaya for the Atlantic lineage. However, Zamora-Silva and Malaquias (Citation2018), based on extended genetic and taxon sampling, supported the validity of Nakamigawaia for both Atlantic N. felis and Pacific N. spiralis and argued that different shell types could be present in the same genus, and considered Migaya a junior synonym of Nakamigawaia.
Specimens from the western Atlantic have consistently been ascribed to the species ‘felis’, but interestingly those from the Pacific have been named erratically by authors and attributed either to the genus Chelidonura or to Nakamigawaia, resulting in pervasive taxonomic confusion and a still unsettled nomenclature, with the name ‘spiralis’, in fact, being hardly used. For example, Susuki (Citation2000, p. 16) and Ono (Citation1999, p. 14, 15) identified specimens from the Izu Peninsula and Kerama Islands (Japan) as Chelidonura sp.; and Gosliner et al. (Citation2008, p. 39) labelled specimens from the Philippines and Papua New Guinea as Nakamigawaia felis, whereas the same authors later used the name Nakamigawaia spiralis (Gosliner et al. Citation2015, p. 45) and Nakamigawaia sp. (Gosliner et al. Citation2018, p. 367). In 2018, Nakano (Citation2018, p. 55; Citation2019, p. 55) identified specimens from Japan as Nakamigawaia felis.
In addition, two distinct chromatic morphs occur in the western Pacific, namely one that is consistent with the original description and having a uniform blackish pattern (Kuroda and Habe Citation1961; Baba Citation1985), and the other characterised by a white-dotted pattern on a brownish background (Ono Citation1999; Gosliner et al. Citation2008, Citation2015, Citation2018; Nakano Citation2018, Citation2019). In this work we use morpho-anatomical characters and a molecular phylogenetic framework to test the taxonomic affiliation of these two morphs and of specimens from the western Atlantic in order to shed light on the diversity and systematics of the genus Nakamigawaia.
Material and methods
Taxon sampling and literature review
Samples were obtained from the collections of the Department of Natural History, University Museum, University of Bergen, Norway (ZMBN). Specimens of Nakamigawaia from Taiwan (two black morph specimens and two white-dotted morph specimens), Japan (black morph), Venezuela and the Bahamas were used for morphological studies, for a total of 11 specimens (six of those sequenced for their DNA; ). In addition, we obtained from GenBank 16 sequences of the cytochrome c oxidase subunit I gene (COI), representing the genera Chelidonura, Nakamigawaia, Philinopsis and Tubulophilinopsis. The tree was rooted with the species Philine quadripartita ().
Table 1. List of specimens used for molecular analysis, including locality, voucher numbers and GenBank accession numbers. *Listed as Aglaja felis in GeneBank; ** listed as Nakamigawaia spiralis in GenBank. Novel sequences generated for this study are depicted in bold font
DNA extraction, amplification and sequencing
DNA was extracted from tissue clipped from the parapodial lobes of the animals following the protocol of the Qiagen DNeasy Blood and Tissue Kit. Sequences of the gene COI were amplified and sequenced using the universal primers by Folmer et al. (Citation1994). Polymerase chain reactions (PCRs) were performed in 25 µL Eppendorf tubes containing 1 μL of DNA, 2.5 μL Qiagen Buffer, 2.5 μL dNTPs, 5 μL Qiagen Q-Solution, 2 μL forward and reverse primers (1 μL for each primer direction), 0.15 μL of Qiagen HotStar TAQ DNA polymerase, 8.35 μL of Sigma water, and 3.5 μL of magnesium chloride. The PCR thermal cycle included an initial denaturation at 95°C for 5 min, followed by 39 cycles of 45 s at 94°C, 45 s at 45°C (annealing temperature), extension at 72°C for 2 min, and a final extension at 72°C for 10 min.
PCRs that did not yield results were redone with the restriction enzyme Takara and the primers GasF1_t1 (TGTAAAACGACGGCCAGTTTTCAACAAACCATAARGATATTGG) and GasR1_t1 (CAGGAAACAGCTATGACACTTCWGGRTGHCCRAARAATCARAA) (Stein et al. Citation2013). In those cases the PCR was started with an initial denaturation step of 5 min at 95°C, followed by 5 cycles of 40 s at 94°C, 40 s at 45°C and 1 min at 72°C, then 35 cycles of 40 s at 94°C, 40 s at 51°C and 1 min at 72°C. This was followed by a final extension step of 5 min at 72°C.
Gel electrophoresis was performed on a 1% agarose/buffer to assess the quality and quantity of the amplified DNA. The successful PCR products were purified with Exonuclease I Shrimp Alkaline Phosphatase (ExoSAP). The total volume of each purification reaction was 10 μL, which consisted of 0.1 μL of Exo I (10 U/μL), 1 μL SAP (1 U/μL), 0.9 μL of Sigma water and 8 μL of PCR product. The mixtures were incubated at 37°C for 30 min, followed by 85°C for 15 min, and finally kept cool at 4°C.
Sanger sequencing reactions were prepared using primers diluted to 3.2 μM. Each sequencing reaction contained 6.5 μL Sigma water, 1 μL buffer, 1 μL BigDye, 1 μL primer and 0.5 μL PCR product (or 1 μL PCR product for samples with a low amount of DNA, with no changes made to the amount of Sigma water). Prior to sequencing the mixtures were run in a thermal cycle including an initial step at 96°C for 5 min, followed by 25 cycles at 96°C for 10 sec, 5 sec at an annealing temperature of 50°C, and 60°C for 4 min, and finally kept cool at 4°C. After the thermal cycle 10 μL Sigma water was added to each sample. Samples were sequenced with an ABI 3730XL DNA Analyser (Applied Biosystems) at the DNA Sequencing Facility, Department of Biological Sciences, University of Bergen, Norway.
Phylogenetic and species delimitation analyses
Both forward and reverse DNA chromatograms were edited and assembled using Geneious R11 (Biomatters, Auckland, New Zealand; Kearse et al. Citation2012). All sequences were blasted using GenBank to test for contamination. Sequences were aligned with the programme Muscle (Edgar Citation2004) implemented in Geneious using default parameters. The alignment was trimmed at both ends where at least 50% of the individual sequences had nucleotides and was translated into amino acids to test for the presence of stop codons. The best-fit model of evolution was estimated with JModelTest 2.1.10 (Darriba et al. Citation2012).
Bayesian molecular phylogenetic inference was done in the program MrBayes 3.2.7 (Ronquist and Huelsenbeck Citation2003) through the platform CIPRES Science Gateway V.3.3 (Phylo.org) with three parallel runs of five million generations with sampling every 100 generations. Node support was assessed using Bayesian posterior probabilities (Felsenstein Citation1985). Convergence of runs was assessed using Tracer 1.7.1 (Rambaut and Drummond Citation2007) with the burn-in set to 25%. The phylogenetic tree was visualised in FigTree 1.4.4 (Rambaut and Drummond Citation2009) and edited in Adobe Illustrator.
Uncorrected p-distances between and within species and colour morphs were estimated with the programme MEGA X (Kumar et al. Citation2018). The Automatic Barcode Gap Discovery method (ABGD; Puillandre et al. Citation2012), implementing the Simple Distance algorithm and default settings, was used to aid in defining candidate species.
Anatomical work and scanning electron microscopy
A total of 11 specimens (three representing the black morph and two the dotted-white morph of N. ‘spiralis’, and six representing N. felis) were dissected for structures of taxonomic relevance, namely the male reproductive system and the shell. Drawings of soft anatomical structures were made under a stereomicroscope with the aid of a camera lucida. Shells were cleansed with diluted commercial bleach, rinsed in distilled water and mounted on metallic stubs covered with carbon sticky tabs, coated with gold–palladium and later studied and imaged with a Fei Quanta 450 scanning electron microscope at the Electron Microscopy Laboratory, Faculty of Mathematics and Natural Sciences, University of Bergen, Norway.
Results
Phylogenetic analyses
The monophyly of the genus Nakamigawia received marginal support in the COI phylogenetic analysis. Three sub-clades were rendered within Nakamigawaia: a clade with Caribbean specimens compatible with the species N. felis, a clade with western Pacific specimens (Taiwan and Lizard Island, Australia) with a mix of black and dotted-white animals that we provisionally assigned to N. ‘spiralis’ (= N. nakanoae sp. nov.; name adopted hereafter; see Taxonomic section), and a single specimen from the Philippines (Nakamigawaia sp.; based on GenBank sequences) that split off alone and probably corresponds to an unnamed species.
The COI genetic uncorrected p-distance between black and white-dotted morphs of N. nakanoae sp. nov. was estimated at 0–0.4%, which – like the phylogenetic results – suggests these two morphs are conspecific. The genetic distance within specimens of N. felis was 0–0.7%, and within specimens of N. nakanoae sp. nov. it was 0–1.2%. The genetic distance between N. felis and N. nakanoae sp. nov. was 18.8–20.1%. The ABGD analysis rendered 11 candidate species including N. felis and N. nakanoae sp. nov. (see Supplementary material).
Taxonomic section
Genus NAKAMIGAWAIA Kuroda and Habe, Citation1961
Nakamigawaia felis (Er. Marcus and Ev. Marcus, Citation1970)(, , , )
Figure 1. Live images of Nakamigawaia nakanoae sp. nov. (A, C, D) and Nakamigawaia felis (B). A, black morph, Taiwan, ZMBN 116778, animal length (L) = 10 mm (paratype). B, the Bahamas, ZMBN 91108, L = 13 mm. C–D, white dotted morph, Taiwan, ZMBN 116777, L = 8 mm (paratype).

Figure 2. Bayesian phylogeny of Aglajidae species with a focus on the genus Nakamigawaia, based on the mitochondrial Cytochrome c oxidase subunit I (COI) gene. Numbers on branches are posterior probabilities. Tree rooted with Philine quadripartita. BM = black morph. DM = white dotted morph.

Diagnosis
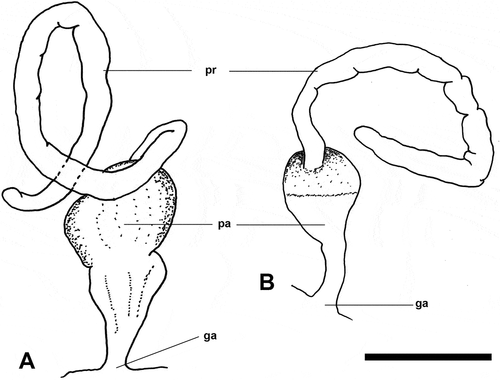
External colouration plain black. Internal shell dilated with an open whorl extending half a turn, protoconch dorsally covered by teleoconch, a plate-like structure can project ventrally near protoconch. Caudal lobes of nearly equal length. Male reproductive system with pyriform penial chamber and tubular, long, folded prostate. Prostate about 3 times longer than penial chamber.
Examined material
Venezuela, Isla Tortuga, Playa El Yaque’s Lagoon, 2 specimens dissected and sequenced, ZMBN 84913, animal length (L) = 13 mm, coll. Manuel Malaquias, 16 March 2010. The Bahamas, Eleuthera Island, Savannah Sound, 4 specimens dissected, ZMBN 91108, L = 10–13 mm, coll. Manuel Malaquias, 14 April 2013.
External morphology (): Plain black including foot. Caudal lobes about the same length.
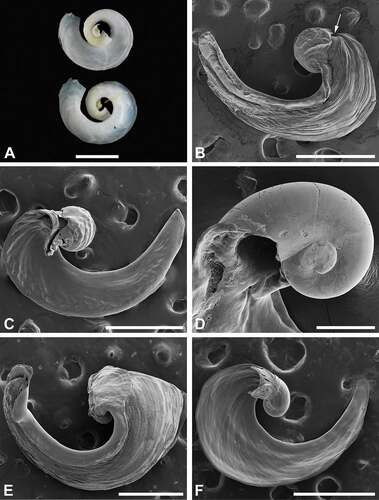
Shell (): Convex, dilated, with an open whorl extending half a turn. Protoconch smooth, partly visible ventrally; dorsally fully covered by extension of teleoconch; a plate-like structure can project ventrally near protoconch (fragile, breaks off easily).
Figure 3. Shells of Nakamigawaia species. A, N. spiralis, holotype, NSMT-39805, ventral (upper) and dorsal (lower) views (macrophotograph), Japan. B, N. nakanoae sp. nov., micrograph of dorsal view; arrow pointing to protoconch, ZMBN 132073, animal length (L) = 7 mm (fixed length), Japan. C, N. nakanoae sp. nov., micrograph of ventral view, ZMBN 132073, L = 7 mm (fixed length), Japan. D, N. nakanoae sp. nov., micrograph of protoconch observed from ventral side of shell, ZMBN 116778, L = 10 mm, Taiwan. E, N. felis, micrograph of dorsal view, ZMBN 91108, L = 13 mm, the Bahamas. F, N. felis, micrograph of ventral view, ZMBN 91108, L = 12 mm, the Bahamas. Scale bars: a = 200 μm; b, c, e, f = 500 μm; d = 100 μm.
Male reproductive system (): Penial chamber pyriform, rounded distally and funnel-like proximally. Length between genital opening and insertion of prostate around 1 mm. Prostate tubular, cylindrical, folded; length around 3 mm.
Ecology
Found crawling on sand between 0.2 and 1 m depth.
Distribution
Caribbean Sea, between the Bahamas in the north and Venezuela in the south (Malaquias Citation2014; Caballer et al. Citation2015).
Nakamigawaia nakanoae Hellem and Malaquias sp. nov.())
Zoobank: lsid:zoobank.org:act:EB686586-9EFD-448A-B743-15C0FDC583DF
Diagnosis
External colouration plain black or brownish dotted in white. Internal shell dilated with an open whorl extending half a turn, protoconch dorsally visible near transition to teleoconch, otherwise covered by teleoconch layer; ventrally a funnel-like structure can be present near protoconch. Right caudal lobe nearly half length of left lobe. Male reproductive system with pyriform penial chamber and tubular, long, folded prostate. Prostate about 5 times longer than penial chamber.
Type locality
Taiwan, Pingtung County, Kenting National Park, Shadao (21.912233N, 120.846961E).
Etymology
The species is named after Dr Rie Nakano, for her passion and contributions to the study of the sea slug molluscs of Japan and for her continuous support throughout the years of our research work.
Examined material
Taiwan, Pingtung County, Kenting National Park, Shadao, 1 specimen, ZMBN 130245 (black morph), L = 10 mm, holotype, coll. Manuel Malaquias, 7 May 2017. Taiwan, Pingtung County, Kenting National Park, Shadao, 2 specimens, ZMBN 116814 (black morph), L = 10 mm, paratypes, coll. Manuel Malaquias, 7 May 2017. Taiwan, Pingtung County, Kenting National Park, Shadao, 2 specimens dissected and sequenced plus 46 observed, ZMBN 116778 (black morph), L = 8–10 mm, paratypes, coll. Manuel Malaquias, 7 May 2017. Taiwan, Pingtung County, Kenting National Park, Shadao, 2 specimens dissected and sequenced plus 5 observed, ZMBN 116777 (white dotted morph), L = 8 mm, paratypes, coll. Manuel Malaquias, 7 May 2017. Japan, Kagoshima, Amami-Ohshima Island, Ayamaru, 1 specimen dissected, ZMBN 132073 (black morph), L = 7 mm (fixed length), coll. Rie Nakano, 19 May 2015.
External morphology ()): Plain black including foot, or brownish background dotted in white, with rim of head-shield and parapodial lobes white or dashed-white; foot sole brownish, white-dotted. Caudal lobes asymmetrical, with right lobe nearly half length of left lobe.
Shell ()): Convex, dilated, with an open whorl extending half a turn. Protoconch smooth, entirely or partly visible ventrally; dorsally visible only near transition to teleoconch; a funnel-like structure (fragile, breaks off easily) can be present ventrally near protoconch.
Male reproductive system ()): Penial chamber pyriform, rounded distally and funnel-like proximally. Length between genital opening and insertion of prostate around 1 mm. Prostate tubular, cylindrical, folded; length around 5 mm. One specimen from Japan (ZMBN 132073) has a 7.5 mm prostate.
Ecology
Specimens were found at depths between 1 and 6 m crawling on white sandy bottoms.
Distribution
Kenting in Taiwan (current study), Kerama Islands, Amami-Ohshima Island and Izu Peninsula in Japan (Ono Citation1999; Susuki Citation2000), the Philippines and Papua New Guinea (Gosliner et al. Citation2008), and in Lizard Island, eastern Australia (current study).
Discussion
There is widespread confusion and inconsistency about the naming of slugs with black-coloured headshields occurring in the western Pacific (Kuroda and Habe Citation1961; Baba Citation1985; Ono Citation1999; Gosliner et al. Citation2008, Citation2015, Citation2018; Nakano Citation2018, Citation2019) and controversy regarding the definition of the genus Nakamigawaia (Ortea et al. Citation2014; Zamora-Silva and Malaquias Citation2018). The original description in Japanese of the type species N. spiralis is brief and includes a single illustration of the shell (). The description refers to animals with black body colour and a shell coiling in three turns, opening downwards. Specimens were collected by Mr Korokuro Nakamigawa at ‘Zushi beach’ [= Kasajima], Kanagawa Prefecture, in a tide pool (Kuroda and Habe Citation1961; Baba Citation1985).
The first comprehensive study of N. spiralis was performed by Baba (Citation1985). The author provided detailed data on the morphology and anatomy and sound evidence of its taxonomic affinity with the family Aglajidae. Striking features of the description by Baba (Citation1985) are the comparatively large size of the animals (ranging between 25 and 35 mm) and the long prostate (about 20 mm for a specimen 25 mm in length overall; measurements based on in Baba Citation1985).
In our preliminary identification of black headshield slugs collected in southern Taiwan and southern Japan we assigned these specimens to N. spiralis. However, detailed comparative studies of the shell and reproductive system revealed substantial differences between what was observed and what is known about these structures in N. spiralis. The shells in our specimens were dilated, with an open whorl extending half a turn (), and there were differences in the soft anatomy: prostates were shorter, ranging between 3.5 and 6.5 mm in specimens that measured about 10 mm in total length when alive. However, the difference in the size of the reproductive system might be explained by the unique length of the animals. Interestingly, such slugs with large black headshields have not been observed or reported during at least the last decade (Rie Nakano, personal communication). Therefore, based on differences in the shell shape and apparent dissimilarities in parts of the male reproductive system, we assign our specimens to a new species, named Nakamigawaia nakanoae sp. nov. The known distribution of N. spiralis from Sagami Bay to the Kyusyu region (Kuroda and Habe Citation1961; Baba Citation1985) suggests a species of temperate affinity, whereas N. nakanoae sp. nov. is confined to tropical waters.
This study confirms the validity of the species N. felis and its tropical western Atlantic geography, and suggests the occurrence of an additional species of Nakamigawaia present in the Philippines at least. No phylogenetic or genetic differentiation was detected between the white-dotted and black morphs of N. nakanoae sp. nov. (COI uncorrected p-distance ranged between 0% and 0.4%), whereas the large genetic difference between the latter species and N. felis (18.8–20.1%) suggests an old speciation event or fast evolutionary rates in these species ().
Externally, N. felis and the black morph of N. nakanoae sp. nov. are nearly indistinguishable, but there is one subtle difference: namely, the right and left caudal lobes in the latter species are of different lengths, whereas in N. felis they are of similar size (). Also, the internal shells of these two species are similar, but in N. felis the dorsal part of the protoconch is covered by a teleoconch layer, whereas in N. nakanoae sp. nov. part of the protoconch is dorsally visible (). The male reproductive systems are also similar in morphology, yet the ratio between the length of the prostate and the length of the penial chamber (from the genital aperture to the insertion point of the prostate) is about 5× in N. nakanoae sp. nov. and only 3× in N. felis ().
The systematics of Nakamigawaia and its recognised lineages (N. spiralis, N. felis, N. nakanoae sp. nov., Nakamigawaia sp. from the Philippines) admittedly remains elusive after this work. Our COI gene tree barely recovers support for the monophyly of the genus, but this is not surprising considering that it is a single-locus phylogeny. On the other hand, the works of Camacho-García et al. (Citation2014) and Zamora-Silva and Malaquias (Citation2018), based on a broader character set, strongly support the monophyly of Nakamigawaia. However, if the genus Nakamigawaia is to be defined by the presence of a spiralled internal shell and a long and convoluted prostate, then perhaps N. felis, N. nakanoae sp. nov. and eventually Nakamigawaia sp. will have to be assigned to a distinct genus. In this case, the junior synonym name Migaya Ortea, Caballer and Espinosa, Citation2014 is available. Nevertheless, answering this question warrants the study of new specimens of the true N. spiralis with spiralled shells and its comparison in a phylogenetic framework with other aglajids including the species currently assigned to Nakamigawaia. Until additional evidence is available, we believe that the use of the generic name Nakamigawaia for these lineages better promotes taxonomic stability.
Acknowledgements
We are very thankful to Rie Nakano in Japan and Kasunori Hasegawa at the National Museum of Nature and Science, Tokyo, for sharing material with us, valuable discussions about type material and type localities, and helping translating Japanese texts; to Chung-Chi Hwan (National University of Kaohsiung, Taiwan) for help organising a fieldtrip to Taiwan and for his friendship; to Trond Oskars (Statsforvalteren at Møre og Romsdal, Norway) for help sampling in Taiwan and companionship; and to the authorities of the Kenting National Park in Taiwan for granting collecting permits and access to laboratory and accommodation facilities. At the University of Bergen, we are indebted to Irene Hegstad (Electron Microscopy Lab, Faculty of Mathematics and Natural Sciences) for help with electron scanning microscopy, and to Louise Lindblom and Justine Siegwald for help with DNA bench work. In addition, we are grateful to the two anonymous referees whose comments and suggestions contributed to a better version of this work.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Baba K. 1985. Anatomical review of a Cephalaspidean mollusc, Nakamigawaia spiralis Kuroda & Habe in Habe 1961, (Aglajidae), from Japan. Spec Publ Mukaishima Mar Biol Stat. 231:1–5.
- Burn R, Thompson TE. 1998. Order Cephalaspidea. In: Beesley GJ, Ross B, Wells AA, editors. Fauna of Australia, Mollusca: the Southern Synthesis. Melbourne: CSIRO Press; p. 943–959.
- Caballer M, Ortea J, Rivero N, Tucker GC, Malaquias MAE, Narciso S. 2015. The opisthobranch gastropods (Mollusca: Heterobranchia) from Venezuela: an annotated and illustrated inventory of species. Zootaxa. 4034:201–256. doi:https://doi.org/10.11646/zootaxa.4034.2.1.
- Camacho-García YE, Ornelas-Gatdula E, Gosliner TM, Valdés Á. 2014. Phylogeny of the family Aglajidae (Pilsbry, 1895) (Heterobranchia: Cephalaspidea) inferred from mtDNA and nDNA. Mol Phylogenet Evol. 71:113–126. doi:https://doi.org/10.1016/j.ympev.2013.11.010.
- Chaban EM, Ekimova IA, Schepetov DM, Kohnert PC, Schrödl M, Chernyshev AV. 2019. Euopisthobranch mollusks of the order Cephalaspidea (Gastropoda: Heterobranchia) of the Kuril-Kamchatka Trench and the adjacent Pacific abyssal plain with descriptions of three new species of the genus Spiraphiline (Philinidae). Prog Oceanogr. 178:102185. doi:https://doi.org/10.1016/j.pocean.2019.102185.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModeltest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772. doi:https://doi.org/10.1038/nmeth.2109.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. doi:https://doi.org/10.1093/nar/gkh340.
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evol. 39:783–791. doi:https://doi.org/10.1111/j.1558-5646.1985.tb00420.x.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3:294–299.
- Gosliner TM. 1980. Systematics and phylogeny of the Aglajidae (Opisthobranchia: Mollusca). Zool J Lin Soc. 68:325–360. doi:https://doi.org/10.1111/j.1096-3642.1980.tb01925.x.
- Gosliner TM, Behrens DW, Valdés Á. 2008. Indo-Pacific nudibranchs and sea slugs: a field guide to the world’s most diverse fauna. Washington: Sea Challengers Natural History Books & California Academy of Sciences.
- Gosliner TM, Behrens DW, Valdés Á. 2015. Nudibranch & Sea Slug identification - Indo-Pacific. Second ed. Jacksonville (Florida, USA): New World Publications Inc.
- Gosliner TM, Behrens DW, Valdés Á. 2018. Nudibranch & Sea Slug identification - Indo-Pacific. Second ed. Jacksonville (Florida, USA): New World Publications Inc.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649. doi:https://doi.org/10.1093/bioinformatics/bts199.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K, Battistuzzi FU. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. doi:https://doi.org/10.1093/molbev/msy096.
- Kuroda T, Habe T. 1961. Nakamigawaia spiralis. In: Habe T, editor. Coloured illustrations of the shells of Japan (II). Vol. 183: p. 66. pls., 42 1-Chome Uehonmachi Higashi-Ku, Osaka, Japan. (In Japanese).
- Malaquias MAE. 2014. New data on the heterobranch gastropods (‘opisthobranchs’) for the Bahamas (tropical western Atlantic Ocean). Mar Biodivers Rec. 7:e27. doi:https://doi.org/10.1017/S175526721400030X.
- Marcus E, Marcus E. 1970. Opisthobranchs from Curaçao and faunistically related regions. Stud Fauna Curaçao Other Caribb Isl. 122:1–129.
- MolluscaBase. eds. (2021). Chelidonuridae Habe. MolluscaBase. 1961. http://marinespecies.org/aphia.php?p=taxdetails&id=22981
- Nakano R. 2018. Field guide to sea slugs and Nudibranches of Japan. Tokyo: Bun-ichi Co., Ltd.
- Nakano R. 2019. Field guide to sea slugs and Nudibranches of Japan. Second ed. Tokyo: Bun-ichi Co., Ltd.
- Ono A. 1999. Opisthobranchs of Kerama Islands. Japan: TBS-Britannica & Co., Ltd.
- Ortea J, Caballer M, Moro L, Espinosa J. 2014. What the shell tells in Aglajidae: a new genus for Aglaja felis (Opisthobranchia: Cephalaspidea). Rev Acad Canar Cienc. 26:83–119.
- Puillandre N, Lambert A, Brouillet S, Achaz G. 2012. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol. 21:1864–1877. doi:https://doi.org/10.1111/j.1365-294X.2011.05239.x.
- Rambaut A, Drummond A, 2009. FigTree v1.4.4. http://tree.bio.ed.ac.uk/software/figtree/
- Rambaut A, Drummond AJ 2007. Tracer v1.7.1. http://tree.bio.ed.ac.uk/software/tracer/
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574. doi:https://doi.org/10.1093/bioinformatics/btg180.
- Rudman WB. 1978. A new species and genus of the Aglajidae and the evolution of the Philinacean opisthobranch molluscs. Zool J Lin Soc. 62:89–107. doi:https://doi.org/10.1111/j.1096-3642.1978.tb00524.x.
- Stein ED, White BP, Mazor RD, Miller PE, Pilgrim EM, Steinke D. 2013. Evaluating ethanol-based sample preservation to facilitate use of DNA barcoding in routine freshwater biomonitoring programs using benthic macroinvertebrates. PLoS One. 8:1–7. doi:https://doi.org/10.1371/journal.pone.0051273.
- Susuki K. 2000. Opisthobranchs of Izu Peninsula. Tokyo (Japan): TBS-Brittanica Co., Ltd.
- Zamora-Silva A, Malaquias MAE. 2016. Diet preferences of the Aglajidae: a family of cephalaspidean gastropod predators on tropical and temperate shores. J Mar Biolog Assoc UK. 96:1101–1112. doi:https://doi.org/10.1017/S0025315415000739.
- Zamora-Silva A, Malaquias MAE. 2018. Molecular phylogeny of the Aglajidae head-shield sea slugs (Heterobranchia: Cephalaspidea): new evolutionary lineages revealed and proposal of a new classification. Zool J Lin Soc. 183:1–51. doi:https://doi.org/10.1093/zoolinnean/zlx064.