ABSTRACT
A new species of Cyrtodactylus is described from Tamenglong District in northwestern Manipur State, northeast India. The new species is diagnosed from other congeners based on a combination of morphological characters and molecular data (NADH dehydrogenase subunit 2 gene, ND2). Molecular analyses placed the new species as the sister taxon to C. montanus. Due to inconsistencies in the hierarchical clade terminology for Cyrtodactylus species in the Indo-Burma region, recommendations are made for stabilising the terminology and the names for various levels of clades. Based on this revised terminology, the new species was found to be nested within the gansi group of the khasiensis clade, within the Indo-Burma radiation of Cyrtodactylus. A large number of errors and inconsistencies in literature covering species of the Indo-Burma radiation were discovered, particularly in summary tables of measurements and meristic data. Other notable errors found included incorrect identifications of type specimens in figure captions, errors in museum catalogue numbers of type series, incorrect GPS coordinates for type localities, a variety of issues involving GenBank accession numbers and associated data, and many others. We highlight these specific errors and inconsistencies and attempt to clarify or correct them where possible. Suggestions and recommendations are given for authors, journal editors and reviewers on how to improve the accuracy and consistency of morphological and molecular data provided in taxonomic and molecular phylogenetic papers. We also encourage authors and scientific journals to follow or enforce the basic citation guidelines set by NCBI when using sequences downloaded from GenBank, which have been largely overlooked, or ignored. It is hoped that the issues discussed here will be taken as a reminder that errors in peer reviewed papers are an inevitability, and that users of this literature must always carefully assess the accuracy of others’ data when it is important for the interpretation of their results to reduce the propagation of errors in the literature.
http://www.zoobank.org/urn:lsid:zoobank.org:pub:9E00496B-C8D0-4391-B1E5-18522B221FBD
Introduction
Cyrtodactylus Gray, Citation1827, is a widely distributed genus of geckos found from northern Australia through Southeast Asia and west across South Asia into northern Pakistan, extending as far north as the Tibetan Plateau (e.g. Wood Jr et al. Citation2012; Bauer et al. Citation2013). Currently, more than 300 species have been described in the genus, a number that is rapidly increasing each year (e.g. Riyanto et al. Citation2020; Grismer et al. Citation2021). Until recently, few taxonomic studies had been carried out on the genus Cyrtodactylus from northeast India with only three species reported between 1870 and 2017, Cyrtodactylus khasiensis (Jerdon, Citation1870), C. himalayicus (Annandale, Citation1906), and C. gubernatoris (Annandale, Citation1913). The latter two species are still only known from two localities each, their respective type localities and one additional nearby locality (Agarwal et al. Citation2018a). Cyrtodactylus khasiensis, however, had been reported from throughout northeast India, Bhutan, Bangladesh, Tibet and western Myanmar (e.g. Smith Citation1935; Ahsan Citation1998; Pawar and Birand Citation2001; Li Citation2007; Agarwal et al. Citation2014; Das et al. Citation2016). Mahony and Reza (Citation2008) and Mahony et al. (Citation2009a, Citation2009b) were the first to question the taxonomic status of populations reported from Bangladesh, and Mahony (Citation2009) reviewed the taxonomic status of those reported from Myanmar. Mahony (Citation2009) and Mahony et al. (Citation2009b) demonstrated that C. khasiensis from these regions was a catch-all name given to multiple morphologically distinct species. Agarwal et al. (Citation2014) sampled populations from across northeast India in a molecular phylogenetic study and provided strong evidence that many deeply divergent and undescribed taxa remained in the C. khasiensis complex requiring further taxonomic attention. Since then, 10 new species have been described from northeast India alone representing many of the genetic lineages identified by Agarwal et al. (Citation2014) from the states of Arunachal Pradesh, Assam, Meghalaya, Nagaland, Tripura, and West Bengal (Agarwal et al. Citation2018a, Citation2018b; Purkayastha et al. Citation2020; Mirza et al. Citation2021). Herein we describe, based on its distinctive morphology and molecular data, the 14th species of Cyrtodactylus from northeast India; this species is the first to be described from Manipur State, which is geographically located in the Indo-Burma Biodiversity Hotspot (Mittermeier et al. Citation2004). The systematic position of the new species is elucidated within the Indo-Burma radiation of Cyrtodactylus based on molecular phylogenetic analyses using the ND2 gene.
A review of the relevant literature on related Cyrtodactylus species has revealed a large and perhaps worrying number of errors. Many of these errors are subtle (e.g. typos), and perhaps easily overlooked, but those involving morphological characters have led to perpetuated misrepresentations that have in turn led to inaccurate diagnoses of subsequent new species. We identify and attempt to correct these errors where possible, and highlight a wide variety of others (e.g. involving GenBank numbers and related data, type locality data, misinterpretation of published phylogenetic analyses, confusions regarding the number of type specimens and their associated registrations, misidentification of type specimens in figures, etc.) in an attempt to mitigate, or at least reduce the propagation of errors. The types and range of errors identified here are very unlikely to be restricted to literature related to this taxonomic group, thus we make suggestions on measures that may be taken by authors, journal editors and reviewers that will reduce the quantity of such errors being published in the future.
Materials and methods
Fieldwork and museum abbreviations
Fieldwork was conducted in Namtiram Village in Tamenglong District, Manipur State, northeast India by Rachunliu G. Kamei (RGK) in May 2007, June 2008 and July 2015. The single specimen of the new species described herein (BNHS 2751) was located during an opportunistic visual herpetofaunal survey. GPS coordinates and elevation for the collection locality were recorded using a Garmin GPSMAP® 62s. The specimen was photographed in life, humanely euthanised by injecting 2% lidocaine solution into the body cavity, fixed in 5% aqueous formalin and washed in water for ca. 24 hours before being transferred to 70% ethanol for long-term preservation. Prior to fixation, muscle tissue from the chest was excised and stored in PCR grade absolute ethanol at −4°C for molecular analyses. Abbreviations for museum collections are: Bernice Pauahi Bishop Museum, Honolulu, Hawaii, USA (BPBM); Bombay Natural History Society, Mumbai, Maharashtra, India (BNHS); Brigham Young University, Provo, Utah, USA (BYU); California Academy of Sciences, San Francisco, California, USA (CAS); Centre for Ecological Sciences, Bangalore, India (CES); Chulalongkorn University Museum of Zoology, Bangkok, Thailand (CUMZ); Field Museum of Natural History, Chicago, Illinois, USA (FMNH); La Sierra University Herpetological Collection, La Sierra University, Riverside, California, USA (LSUHC); Muséum d’histoire naturelle de Genève, Geneva, Switzerland (MHNG); Museum of Vertebrate Zoology, Berkeley, California, USA (MVZ); Departmental Museum of Zoology, Mizoram University, Aizawl, Mizoram, India (MZMU); Natural History Museum, United Kingdom, London, UK (NHMUK; previously British Museum [Natural History], BMNH); National Museum of Natural History, Smithsonian, Washington, D.C., USA (NMNH; previously United States National Museum, USNM); North East Regional Centre, Zoological Survey of India, Shillong, Meghalaya, India (NERC/ZSI; previously the Eastern Regional Station, Zoological Survey of India, ERS/ZSI); Pakistan Museum of Natural History, Islamabad, Islamabad Capital Territory, Pakistan (PMNH); Zoological Survey of India, Kolkata, West Bengal, India (ZSIK).
Molecular analyses
The tissue sample for BNHS 2751 was digested and genomic DNA extracted using a DNeasy Blood and Tissue Kit (Qiagen) following manufacturer’s instructions. The complete ND2 gene was targeted for amplification based on the availability of homologous sequences for Cyrtodactylus on GenBank (Benson et al. Citation2017). The primers, L4437b (forward) and H5540 (reverse) (Macey et al. Citation1997) were used for PCR amplification and sequencing. The PCR was performed in a 25 μl reaction composed of 1.5 μl extracted DNA (ca. 10 ng/μl), 10 μl PCR grade H2O, 12.5 μl MyTaq™ Mix (Bioline) and 0.5 μl each of forward and reverse primers (10 ng/μl). The PCR reaction protocol involved the following steps: initial denaturation at 95°C for two minutes, denaturation for 40 cycles at 95°C for 30 seconds, annealing at 53°C for 30 seconds, and extension at 72°C for one minute. The final extension was at 72°C for two minutes. PCR purification and Sanger sequencing (forward and reverse) were outsourced to the genomics services company Medauxin (Bangalore, India). Chromatograms for the forward and reverse sequences were checked for quality and assembled in Geneious V.8.1.9 (Kearse et al. Citation2012). A BLASTn (Altschul et al. Citation1990) search was performed for the sequence on the NCBI BLAST website (http://blast.ncbi.nlm.nih.gov) to confirm genus-level identity against GenBank sequences. Sequences for all known species (on 28 January 2021) and divergent unnamed lineages within the Indo-Burma clade (sensu Agarwal et al. Citation2014) and representative outgroup taxa of Cyrtodactylus were downloaded from GenBank. Several outgroup combinations were experimented with until the topology of the Indo-Burma clade (sensu Agarwal et al. Citation2014) was consistent between analyses, and resembled phylogenies obtained elsewhere that were considered more reproduceable (/‘reliable’) since they were based on more loci and included more outgroups than our study (e.g. Agarwal et al. Citation2014). Outgroup optimisation was used to mitigate introducing potential bias/instability in our final tree from unintentional inappropriate outgroup selection (e.g. a ‘rogue taxa’). Sequences used in the finalised alignment were generated by the following studies: Agarwal et al. (Citation2014, Citation2018c); Bauer et al. (Citation2013); Grismer et al. (Citation2018a, Citation2018b, Citation2019a, Citation2019b); Johnson et al. (Citation2012); Muansanga et al. (Citation2020); Purkayastha et al. (Citation2020); Siler et al. (Citation2010); Wood Jr et al. (Citation2012); the exception being one sequence for C. myaleiktaung Grismer, Wood, Thura, Win, Grismer, Trueblood and Quah, 2018, which at the time of writing was not available on GenBank (see point no. 19 in ‘Errors in literature’ section). This C. myaleiktaung sequence was provided by the lead author of the original publication (Grismer pers. comm. 2020). A full list of sequences is given in . Sequences were aligned in MEGA7 (Tamura and Nei Citation1993; Kumar et al. Citation2016) using the MUSCLE algorithm (Edgar Citation2004), adjusted for open reading frame and inspected by eye for premature stop codons. Non-coding regions were adjusted by eye for obvious alignment errors. Phylogenetic relationships were estimated using RAxML-HPC2 (Stamatakis Citation2014) on XSEDE (CIPRES platform: Miller et al. Citation2010) for maximum likelihood (ML), on an unpartitioned alignment dataset using the GTR GAMMA model under default settings for 1000 bootstrap (bs.) replicates. The tree was visualised in FigTree (Rambaut Citation2009) and C. tibetanus (Boulenger, Citation1905) was selected to root the tree in accordance with results obtained in broader phylogenetic studies of Cyrtodactylus (e.g. Wood Jr et al. Citation2012; Agarwal et al. Citation2014).
Table 1. List of Cyrtodactylus ND2 sequences used in this study (newly generated sequence in bold)
Morphology
Measurements were made using Mitutoyo™ dial callipers to the nearest 0.1 mm. Measurements and scale counts were made on the right side of the specimen unless otherwise stated. The following abbreviations and terminology are used for morphometric measurements and meristics: SVL, snout to vent length; TRL, trunk length, between the axilla and groin; BW, maximum body width; TL, tail length, from the cloaca to the tail tip; TW, maximum tail width; TD, maximum tail depth; HL, head length from the snout tip to the retroarticular process of the jaw; HW, maximum head width; JW, jaw width, taken ventrally at the retroarticular process of the jaw; HD, maximum head depth; FL, forearm length, taken from the elbow to the wrist; CL, crus length, taken from the knee to the base of the foot (i.e. portion of the hindlimb that contains the tibia); OD, orbit diameter, taken between the anterior and posterior bony orbital borders (not comparable with eyeball diameter used in some other studies since this is considered less reliable due to deformation based on the level of hydration of specimens); NO, distance between the posterior edge of the nostril and the anterior bony orbital border; SO, distance between the snout tip and the anterior bony orbital border; OE, distance between the posterior bony orbital border and the anterior border of the ear; EL, maximum ear length/diameter; ES, anterior border of the ear to the snout tip; IN, internarial distance; IO, minimum interorbital distance between left and right supraciliary rows; FW, minimum frontal width; RD, maximum rostral depth; RW, maximum rostral width; ML, maximum mental length; MW, maximum mental width; PMIL, maximum length of the inner postmental; PMOL, maximum length of the outer postmental; FIVL, length of digit IV of the manus, measured from the apex with digit III to the tip of digit IV (excluding the claw); TIVL, length of digit IV of the pes, measured from the apex with digit III to the tip of digit IV (excluding the claw); PcP, precloacal pores, a continuous series of pore-bearing scales on the precloacal region that does not extend onto the thighs; PcFP, precloacofemoral pores, a continuous series of pore-bearing scales that extends from the precloacal region onto the thighs; MVSR, mid-ventral scale rows, counted between ventrolateral folds; PVT, paravertebral tubercles on the trunk only, counted between the level of the axilla and the level of the groin (not comparable with PVT as used in some other studies that count from the occipital region to the mid sacrum); DTR, dorsal tubercle rows, counted transversely across the body at its widest point; SL, total number of supralabials. Two separate counts for subdigital lamellae were recorded on all digits of the right manus and right pes, a count for the basal series, that includes scales of a width at least twice the diameter of palmar and plantar scales up to and including a single large scale at the digital inflection, and a count for the apical series, comprising lamellae distal to the digital inflection but not including the ventral claw sheath and the small nonlamellar scales between the basal and apical lamellae series (counted separately), abbreviations as follows: on the manus, FILam, FIILam, FIIILam, FIVLam, FVLam; on the pes, TILam, TIILam, TIIILam, TIVLam, TVLam.
The new species is morphologically compared with all members of the ‘Mountain clade’ of Cyrtodactylus (sensu Agarwal et al. Citation2014), along with regional congeners that have not yet been subjected to molecular analyses. Characters for the taxa compared to our new species have been obtained from original descriptions, or papers that have included type specimens of relevant taxa for morphological redescriptions. Citations are provided for the source of each character used in the comparisons section. See Appendix I for a list of comparative specimens examined for this study. Herein, the morphological comparison section is to be considered a diagnosis for the newly described species. We have intentionally omitted a separate ‘Diagnosis’ section (that traditionally lists morphological characters assumed to define a given species) for several reasons, one of which, is that the newly described species is presently known from a single specimen. Such a section would be premature, and immediately outdated as further new species are described from the group, and/or when known morphological variation is expanded based on new material of this or related species. Outside of taxonomic revisions of well-studied species groups, species ‘Diagnoses’ are quickly outdated and rarely updated. It is our opinion that including a section entitled so definitively as a species ‘Diagnosis’ would thus be deceptive to readers who do not remain fully up-to-date on relevant literature, or who are unaware of the transient accuracy of species ‘Diagnoses’ in taxonomic groups that contain many poorly defined or yet to be described taxa.
Map
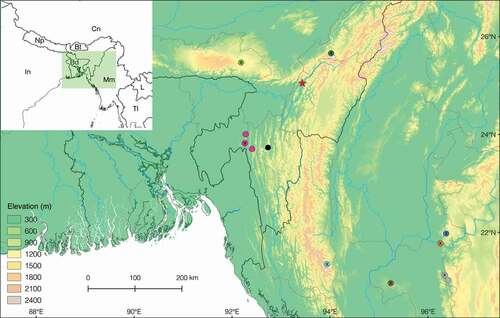
GPS coordinates of localities depicted on the distribution map () were taken from the relevant literature and converted to decimal degrees format using GPS Visualiser (https://www.gpsvisualizer.com/calculators) (Appendix II). The topographic map was made in Quantum GIS (QGIS v.2.14.3-Essen) using the 250 m spatial resolution Shuttle Radar Topography Mission (SRTM) layer available from DIVA-GIS (http://www.diva-gis.org), and other basic layers from the Natural Earth Quick Start Kit (http://www.naturalearthdata.com).
Figure 1. Topographic map of northeast India and surrounding countries showing the type locality (red star) of Cyrtodactylus namtiram sp. nov. and all known localities for the other members of the gansi group from the following literature: Agarwal et al. (Citation2018b); Bauer (Citation2002, Citation2003); Grismer et al. (Citation2018c); Muansanga et al. (Citation2020). Circle with an x, type localities; without an x, other published localities: C. aunglini (blue), C. brevidactylus (brown), C. chrysopylos (grey), C. gansi (turquoise), C. jaintiaensis (light green), C. montanus (pink), C. nagalandensis (dark green), C. myaleiktaung (orange), Mizoram sp. (black). Country borders black; state borders brown; major rivers blue. Inset: Map of northeast India showing country borders in black, and green box representing the area depicted in the main map; abbreviations: In India, Np Nepal, Bd Bangladesh, Bt Bhutan, Cn China, Mm Myanmar, Tl Thailand, L Laos.
Results and discussion
Phylogeny and systematics
There are currently several alternative non-Linnean species group/clade naming schemes proposed by different authors for discussing the various levels of Cyrtodactylus clades from the Indo-Burma region (Agarwal et al. Citation2014; Grismer et al. Citation2018c, Citation2019b, Citation2021; Purkayastha et al. Citation2020), with little consensus between studies regarding their nomenclature. We herein provide a recommendation for stabilising the terminology for the various levels of phylogenetic clades in an effort to mitigate confusion and to enable us to clearly discuss the results of our analyses. Whenever possible, we favour adapting or utilising the terminology suggested by previous authors, and based on this, suggest a three-tier taxonomic hierarchy for discussing the systematics of the Indo-Burma clade of Cyrtodactylus (): Tier 1: one geographic radiation: ‘Indo-Burma radiation’ (equivalent to the ‘Indo-Burma clade’ Agarwal et al. Citation2014); Tier 2: four species clades that represent the major subclades within the Indo-Burma radiation that each currently comprise one or more species groups: ‘fasciolatus clade’ (equivalent to the ‘fasciolatus group’ sensu Grismer et al. Citation2021), ‘khasiensis clade’ (equivalent to the ‘gansi group’ sensu Grismer et al. Citation2019b, and the ‘khasiensis group’ of Grismer et al. Citation2021), ‘peguensis clade’ (mostly equivalent to the ‘peguensis group’ of Grismer et al. Citation2021, but excluding the clade containing C. slowinskii and C. russelli), and the ‘slowinskii clade’; Tier 3: species groups from within the khasiensis and peguensis clades defined above that are divisible into smaller units (the fasciolatus and slowinskii clades currently do not contain species groups): 1. khasiensis clade contains four species groups: ‘arunachalensis group’ (currently monospecific), ‘gansi group’ (equivalent to the ‘Mountain clade’ sensu Agarwal et al. Citation2014, and the ‘gansi group’ sensu Grismer et al. Citation2018c), ‘khasiensis group’ (equivalent to the ‘Lowland clade’ sensu Agarwal et al. Citation2014, and the ‘khasiensis group’ sensu Purkayastha et al. Citation2020, but not the ‘khasiensis group’ sensu Grismer et al. Citation2021) and the ‘mombergi group’; 2. peguensis clade contains two species groups: ‘gubernatoris group’ and ‘peguensis group’ (the latter equivalent to the peguensis group sensu Grismer et al. Citation2018d and Citation2019a, but not the ‘peguensis group’ sensu Grismer et al. Citation2021). Hereafter, we refer to the various clades using this revised hierarchy and terminology.
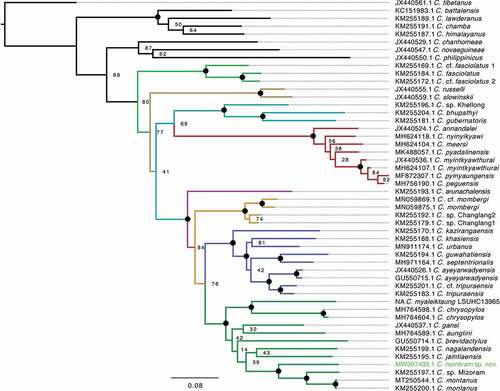
Figure 2. Maximum Likelihood phylogeny of the Indo-Burma radiation of Cyrtodactylus based on the complete ND2, tRNA-Trp, tRNA-Ala genes and partial tRNA-Asn gene alignment. Coloured portion of the tree denotes the Indo-Burma radiation, with species clades and species groups coloured as follows: outgroups (black); fasciolatus clade (light green); slowinskii clade (brown); peguensis clade comprising the gubernatoris group (light blue) and peguensis group (red); khasiensis clade comprising the arunachalensis group (pink), mombergi group (orange), khasiensis group (dark blue), and gansi group (dark green). Bootstrap support values ≥95 represented by a black spot, values <95 are given next to nodes.
Refer to for all known species (described up until 28 January 2021) that can be included within the various species ‘groups’, ‘clades’ and the Indo-Burma radiation based on available DNA sequence data. Grismer et al. (Citation2018c) provisionally proposed the inclusion of Cyrtodactylus mandalayensis Mahony, Citation2009, in the gansi group based on the location of its type locality and supposed morphological similarity to C. aunglini Grismer, Wood, Thura, Win, Grismer, Trueblood and Quah, 2018c, and Grismer et al. (Citation2019b) included C. tamaiensis (Smith, Citation1940) in their ‘gansi group’ without providing any justification. We recommend not restricting the placement of C. mandalayensis and C. tamaiensis to any specific clade, but consider them as incertae sedis within the Indo-Burma radiation of Cyrtodactylus along with the following additional species described from the region that have also not yet been included in a molecular phylogenetic study: C. cayuensis Li, Citation2007, C. himalayicus (Annandale, Citation1906), C. markuscombaii (Darevsky, Helfenberger, Orlov and Shah, Citation1998), and C. martinstollii (Darevsky, Helfenberger, Orlov and Shah, Citation1998). Until their systematic positions within Cyrtodactylus have been determined, all six of the aforementioned species must also be included in the morphological comparison of taxa described as new species in future that are determined or suspected to be members of the Indo-Burma radiation.
The newly generated sequence for the specimen of our new species (BNHS 2751) comprised most of the ND2 gene after editing (968 bp in length out of total 1035 bp). The overall alignment length used for phylogenetic analyses was 1307 bp in length and included the complete ND2, tRNA-Trp, tRNA-Ala genes, and partial tRNA-Asn gene, which were available for most of the other sequences used in the alignment. BNHS 2751 is recovered as a member of the gansi group in the khasiensis clade of the Indo-Burma radiation (), and was resolved as the sister taxon to a clade comprising Cyrtodactylus montanus Agarwal, Mahony, Giri, Chaitanya and Bauer, 2018 and an unnamed taxon from Mizoram State, northeast India (sequenced in Agarwal et al. Citation2014).
The overall phylogeny () generally agreed in topology with previously published phylogenetic trees of the Indo-Burma radiation (e.g. Agarwal et al. Citation2014; Grismer et al. Citation2019b; Purkayastha et al. Citation2020; Mirza et al. Citation2021), with the following exceptions: 1) similar to Grismer et al. (Citation2021: mtDNA dataset, , & mtDNA+nuDNA dataset, ), Mirza et al. (Citation2021: mtDNA dataset, fig. 6), and Purkayastha et al. (Citation2020: mtDNA dataset, ), we recovered the fasciolatus clade as sister to a clade comprising all remaining Indo-Burma radiation taxa, whereas alternatively other studies recovered the slowinskii clade in this position (e.g. Agarwal et al. Citation2014: mtDNA+nuDNA dataset; Grismer et al. Citation2021: nuDNA dataset, ); 2) relationships between consistently unresolved nodes within the gansi group vary between studies (e.g. Agarwal et al. Citation2014; Grismer et al. Citation2018c; Mirza et al. Citation2021; ); 3) relationships between taxa in the peguensis group differ from Grismer et al. (Citation2018d, Citation2019a), likely due to reduced sampling of this group in our analyses.
In contrast to the aforementioned studies, our phylogeny does not resemble those presented in Grismer et al. (Citation2019a, Citation2020) for the Indo-Burma radiation. The phylogeny in Grismer et al. (Citation2019a) portrayed a very different topology for the Indo-Burma radiation to others published (e.g. Grismer et al. Citation2019b), including six species clades instead of the four recovered elsewhere. Unfortunately, the authors neither provided a version of the tree that shows the identity of the sequences in the additional two clades nor did they give a table of sequences used in the study, instead directing the reader to several papers that published partial tables of sequences used in those studies, making it very difficult to duplicate their analyses. Grismer et al. (Citation2019a) did not discuss the significant differences in topology of the Indo-Burma radiation on their tree. The phylogeny in Grismer et al. (Citation2020) despite having been claimed to be ‘nearly perfectly congruent’ with the phylogenies recovered by Wood Jr et al. (Citation2012) and Agarwal et al. (Citation2014), differed from these by showing unusual placements of several taxa within the Indo-Burma radiation, e.g. 1) C. nagalandensis Agarwal, Mahony, Giri, Chaitanya and Bauer, 2018 (gansi group) was nested within the khasiensis group; 2) C. jaintiaensis Agarwal, Mahony, Giri, Chaitanya and Bauer, 2018 (gansi group), a species from India erroneously highlighted as a Burmese taxon, was sister to a clade comprising the mombergi, gansi and khasiensis groups; 3) C. ‘fasciolata’ (sic. for fasciolatus) formed two deeply divergent lineages, one sister to C. tibetanus and the other sister to a clade comprising the remaining Indo-Burma radiation. However, all available ND2 sequences on GenBank consistently formed a single fasciolatus clade within the Indo-Burma radiation comprising relatively closely related lineages in all other studies (e.g. Agarwal et al. Citation2014; Grismer et al. Citation2021; Mirza et al. Citation2021 [‘fasciolatum’ sic. for fasciolatus]; Purkayastha et al. Citation2020; results obtained herein, ). As in Grismer et al. (Citation2019a), Grismer et al. (Citation2020) did not provide a table of sequences they used in their analyses and the disparity in their topology from those obtained in other studies point to possible issues with their analyses. Similar issues are apparent in the topology of the clade that is supposed to represent the Indo-Burma radiation in Grismer et al. (Citation2018c), which differed from all of the aforementioned studies in topology, despite the authors stating that they used a similar sequence dataset and similar analytical techniques. Although the phylogenetic analyses in these studies (Grismer et al. Citation2018c, Citation2019a, Citation2019b) are less focused on resolving the broader phylogeny of Cyrtodactylus but intend to place taxa of interest into clades, outgroup inclusion and topology can have a significant influence on ingroup topology and node support values. For reviewers, editors, and readers who need to evaluate exactly how an analysis has been performed, the task is exponentially more difficult when they are directed to multiple papers for partial lists of raw data. Summarised figures of phylogenies that do not present all of the data portrayed should consistently be supplemented with an original version of the tree and a complete table of sequences used in analyses either within the main body of a paper, or in appendices or supplementary/supporting files.
Taxonomy
Based on the relatively deep genetic distance () between the newly collected Manipur specimen (BNHS 2751) and its closest relatives, and morphological evidence (presented below), we recognise it as a unique, previously unnamed species-level taxon. Despite only a single specimen being available for study, we justify the description of a new species based on this voucher for the following reasons: 1) the specimen is sufficiently morphologically diagnosable against all related taxa; 2) morphologically diagnostic new species of Cyrtodactylus are regularly described based on a single specimen (e.g. C. aequalis Bauer, Citation2003, C. chrysopylos, Citation2003, C. guwahatiensis Agarwal, Mahony, Giri, Chaitanya and Bauer, Citation2018, C. mandalayensis, C. meersi Grismer, Wood, Quah, Murdoch, Grismer, Herr, Espinoza, Brown and Lin, 2018, C. myaleiktaung, C. nyinyikyawi Grismer, Wood, Thura, Win and Quah, 2019; Agarwal et al. Citation2018b; Bauer Citation2003; Grismer et al. Citation2018c; Citation2018d, Citation2019a; Mahony Citation2009); 3) the species appears to occur in low density since three visits were made to the type locality and apparently suitable habitat was surveyed over several nights, but only a single specimen was found; 4) the species was found outside of the region’s protected area network, so it may be under threat from anthropogenic habitat modification. The formal description of the species is hoped to encourage efforts by herpetologists to extend surveys in the region to assess potential conservation threats to this gecko.
Cyrtodactylus namtiram Kamei and Mahony, sp. nov.
Figure 3. Cyrtodactylus namtiram sp. nov. adult male holotype (BNHS 2751) in preservation: (A). dorsal view; (B). ventral view; (C). details of dorsal pholidosis; (D). dorsal view of head; (E). ventral view of head; (F). lateral view of head. (A and B) to scale; (D–F) to scale. Scale bars = 10 mm.

Figure 4. Cyrtodactylus namtiram sp. nov. adult male holotype (BNHS 2751) in life from Tamenglong District, Nagaland State, northeast India: (A). dorsolateral view; (B). close-up of head, left side; (C). ventral view while under anaesthesia, scale bar = 10 mm. Photos taken ex-situ. (D). habitat at the type locality.

Holotype
Adult male, BNHS 2751 (field tag number: RGK 0831), from the vicinity of a waterfall (25.046381, 93.429483, 770 m asl.), Namtiram (ca. 7 km before reaching Aziuram Village coming from Tamenglong Town), Tamenglong West Subdivision, Tousem Tribal Development Block, Tamenglong District, Manipur State, northeast India, collected by Rachunliu G. Kamei on 4 July 2015.
Etymology
The specific epithet is a toponym for the type locality of the species and is treated as a non-Latin noun in apposition. Northeast India witnessed continuous declines in forest and forest habitats due to various factors, but the rural communities are gradually becoming biodiversity conservation-oriented. Local communities are increasingly seeing their forests as a commodity with long-term value worth protecting. We name this species for its type locality to demonstrate the value of Namtiram’s forests as an important refuge for Manipur’s poorly explored biodiversity. We hope that naming the species after the Village will help promote a sense of ownership, pride, and relatability for the local community.
Suggested common name
Namtiram Bent-toed Gecko.
Description of holotype
Holotype (SVL 65.8 mm) in generally good preservation condition. Large portion of skin and muscle tissue removed from right pectoral region as tissue voucher ().
Head moderately long (HL 18.2 mm; HL/SVL 0.28), dorsoventrally depressed (HW 12.0 mm; HD 6.9 mm; HD/HW 0.58), distinct from neck, occipital region slightly enlarged relative to jaw width (JW 10.9 mm; JW/HW 0.91); loreal region convex, interorbital area flat, canthus rostralis blunt, bulbous, forming narrow longitudinal furrow medially on dorsal surface of snout; snout short (SO 7.0 mm; SO/HL 0.38), longer than orbit diameter (OD/SO 0.81); scales on dorsal surfaces of head, eyelids (excluding supraciliaries) and snout primarily homogeneous, granular, longitudinally oval to circular, juxtaposed, those on snout largest; granular scales on occipital region intermixed with larger, rounded granular tubercles, tubercles smallest and sparse dorsally, increasing in size and density laterally over occipital region and posteriorly onto nape where they are two to three times larger than adjacent granules, enlarged tubercles absent on parietal region and remaining surfaces of head. Orbit moderately large (OD 5.7 mm; OD/HL 0.31; IO 6.0 mm; FW 2.7 mm); pupils vertical with crenulated margins; supraciliaries large, mucronate, giving distinctly serrated appearance to eyelids when viewed from above, decreasing in size towards posterior and anterior end of orbits, largest about one-third distance from anterior edge of orbits; enlarged granular scales on upper eyelids absent, tubercle row on outer upper eyelids absent; ear opening oval, obliquely orientated, small (EL 1.2 mm; EL/HL 0.07); orbit to ear distance (OE 4.9 mm) less than orbit diameter (OD 5.7 mm; OE/OD 0.86). Rostral wider (RW 2.9 mm) than deep (RD 1.6 mm), divided dorsally to ca. half rostral depth by well-developed rostral groove; single enlarged supranasal on either side, supranasals separated from each other by three small granular internasals; rostral in contact with first supralabials, nasals, supranasals and three internasals; nostrils oval (NO 5.4 mm; IN 2.3 mm), opening directed postero-laterally, each nasal in broad contact with rostral and surrounded by supranasal, first supralabial, and four small postnasals; mental wider (MW 2.8 mm) than long (ML 2.1 mm), triangular, two well-developed postmentals on either side, inner pair in broad contact behind mental, nearly twice size (PMIL 1.9 mm) of and separating outer pair (PMOL 1.0 mm); each inner postmental bordered by mental, first infralabial, outer postmental and three enlarged gular scales; each outer postmental bordered by inner postmental, first and second infralabials, and four or five enlarged gular scales of dissimilar sizes; supralabials (to midorbital position), nine/eight (left/right); 10 supralabials to angle of jaw on each side; 10 infralabials on each side, second to fourth infralabials bordered by one or two rows of enlarged scales, increasing in size anteriorly; gular scales mostly small, granular, smooth, juxtaposed, homogeneous in size except for those bordering postmentals and where they increase in size posteriorly on throat to become more imbricate.
Habitus slender (BW 13.9 mm; BW/SVL 0.21; TRL 28.1 mm; TRL/SVL 0.43), dorsoventrally depressed; dorsal scales heterogeneous, granular scales mostly rounded, intermixed with irregularly arranged, rounded to bluntly conical enlarged tubercles with a weak keel throughout, becoming more conical and slightly smaller towards lower flanks; dorsal tubercles extend from occipital region posteriorly onto tail base; tubercles on nape smaller than those of dorsum, largest on posterior dorsolateral region, four times larger than adjacent granular scales; ca. 21 dorsal tubercles across mid-dorsum; ca. 33 paravertebral tubercles between axilla and groin; enlarged tubercles on ventrolateral folds absent, ventrolateral folds very weakly developed; ventral scales much larger than dorsal scales, smooth, cycloid, imbricate to subimbricate; ca. 36 ventral scales between ventrolateral folds at mid-trunk; 12 distinct pore-bearing precloacal scales in a continuous ‘V’-shaped series, pore-bearing scales slightly larger than anteriorly contacting ventral scales; one row of enlarged scales between precloacal pore-bearing scales and vent, largest at apex of pore-bearing scale series, approximately two times size of adjacent pore-bearing scales; precloacal groove absent; hemipenal bulge distinct, hemipenes partially everted on both sides (description not provided due to incomplete eversion).
Fore and hindlimbs slender (FL 9.1 mm; FL/SVL 0.14; CL 11.2 mm; CL/SVL 0.17); digits strongly inflected at each joint, all bearing robust recurved claws; enlarged subdigital lamellae not scansorial, lamellar formula on right manus given as proximal (nonlamellar) distal counts: FILam 3(2)6, FIILam 5(2)8, FIIILam 4(1)10, FIVLam 5(2)9, FVLam 4(3)7; proximal (nonlamellar) distal lamellar formula on right pes: TILam 3(3)6, TIILam 5(4)7, TIIILam 6(0)13, TIVLam 7(3)10, TVLam 5(2)10; inter-digital webbing absent on manus and pes; relative length of digits: I < V = II < III < IV (right manus; FIVL 6.0 mm) and I < II < III < V < IV (right pes; TIVL 7.5 mm); scales on palms smooth, flat, juxtaposed to subimbricate; scales on soles smooth, granular, juxtaposed; scales on forelimbs mostly homogeneous, on dorsal and ventral upper arms and ventral forearms covered with small granular subimbricate scales, scales intermixed with few scattered, slightly enlarged blunt tubercles on dorsum of forearms; scales on hindlimbs heterogeneous, dorsal surface of thighs and shanks with small, blunt granular scales intermixed with scattered, enlarged, bluntly conical, weakly keeled tubercles, equally dense on shanks and thighs; preaxial portion of thighs and ventral surface of hindlimbs with enlarged, smooth, flat, imbricate scales slightly smaller in size than (trunk) ventrals, these imbricate scales gradually decrease in size laterally over thighs to become small granular scales on postaxial surface of thighs; femoral pores and enlarged femoral-scale series absent.
Tail complete, original (TL 79.5 mm), slightly dorsoventrally depressed (TD 4.5 mm; TW 5.0 mm; TD/TW 0.90), slender, tapering; caudal segments indistinct; dorsally a single row of enlarged, flat, weakly keeled tubercles positioned transversely on distal edge of each of first three segments of tail (four tubercles on first segment, two each on second and third segments), all other dorsal scales on tail smooth, flat, heterogeneous in size and shape, subimbricate, smallest dorsally but becoming larger on lateral surface, second tail segment with seven rows of middorsal scales; subcaudal scales distinctly larger than dorsal caudal scales, but number of subcaudals per segment could not be assessed as segments are indistinct ventrally, subcaudals smooth, imbricate and heterogeneous in size, no distinct series of enlarged plates or series of enlarged paired subcaudals; four/three (left/right) smooth, bluntly conical post cloacal spurs on each side of tail base.
Colouration in life (): Dorsal and lateral surfaces of head primarily light greyish-brown, a faint brown transverse curved stripe extending from posterior-most edge of eyelids and a short straight faint brown longitudinal stripe on posterior dorsum of head, combined forming a vaguely trident-like marking; a faint brown stripe extends from posterior border of orbit through ear onto side of neck on each side; distinct nuchal collar absent; pale yellowish-brown labials; supraciliaries yellowish-brown; iris pale bluish-grey with a brown venous pattern, pupil with a light-brown border; dorsal and lateral surface of neck and trunk primarily light greyish-brown; seven distinct parallel pairs of dark brown blotches from level of forelimbs to sacrum, anteriorly dark brown blotches fused into a pair of parallel brown longitudinal stripes from posterior edge of head to level of forelimb on right side or beyond level of forelimb on left side, would form approximately nine pairs of blotches if anterior blotches were not fused (fusion/separation of anterior blotches is expected to be variable in this species as observed in other related taxa, e.g. C. montanus, ); dorsal blotches with distinct dark posterior margins, fading anteriorly with indistinct anterior margins; flanks with dark brown and grey blotches; dorsal and lateral surfaces of forelimbs and hindlimbs mottled light grey and brown; tail, dorsally and laterally with 12 contrasting dark brown and light grey transverse bands, light bands slightly narrower than dark bands; entire ventral surface of head, body and limbs white with yellow tinge, more so posteriorly; ventral surfaces of manus and pes light greyish-brown; ventral surface of tail primarily yellow with brown speckling, becoming darker distally where dark and light bands are distinct. Colouration in preservative (): generally similar to colouration in life though less vibrant; yellow tones on supraciliaries and labials became light brown; ventrally pale brownish-cream slightly darker on abdomen; ventral surface of tail pale grey, ventral surfaces of manus and pes pale grey; iris dark grey.
Figure 5. Cyrtodactylus montanus in life showing variation of dorsal markings and colouration. Photos taken in-situ of six different uncollected adult individuals from the Jampui Hills, North District, Tripura State, northeast India: (A). adult female from Phuldungsei Village, nearby the type locality of C. montanus; (B–f). from the vicinity of Vanghmun Village: (B and C). adult females; (D–F). adult males.

Morphological comparisons
Cyrtodactylus namtiram sp. nov. is herein compared to the following species confirmed to be members of the gansi group based on molecular phylogenetic analyses (): C. aunglini; C. brevidactylus Bauer, Citation2002; C. chrysopylos; C. gansi Bauer, Citation2003; C. jaintiaensis; C. montanus; C. myaleiktaung; C. nagalandensis. The new species is further compared with the following Cyrtodactylus species from northeast India and surrounding regions that have not yet been assigned to any clade due to the absence of available molecular data: C. cayuensis; C. himalayicus; C. mandalayensis; C. markuscombaii; C. martinstollii; C. tamaiensis. Characters with conflicting information in the literature that are clarified in the ‘Errors in literature’ section (below) are marked with an asterisk followed by the point number in square brackets.
Cyrtodactylus namtiram sp. nov. differs from C. aunglini by PVT ca. 33 between axilla and groin (vs. between ‘limb insertions’ 37–45*[2], N = 10; Grismer et al. Citation2018c: table 5), dorsal markings on trunk comprise paired dark brown blotches (vs. thin dark brown transverse stripes; Grismer et al. Citation2018c), MVSR ca. 36 (vs. ‘ventral scales’ 41–47*[1], N = 10; Grismer et al. Citation2018c: tables 4 & 5), light coloured caudal bands 12 (vs. 9–11, N = 4; Grismer et al. Citation2018c), nape band absent (vs. present; Grismer et al. Citation2018c); from C. brevidactylus by PVT ca. 33 between axilla and groin (vs. 38–42 between ‘limb insertions’, N = 2; Grismer et al. Citation2018c), PcP 12 (vs. 8–9, N = 2 [or N = 3, if both LSUHC 13498–99 cited in Grismer et al. Citation2018c are males; the sex of these specimens was not explicitly stated]; Bauer Citation2002; Grismer et al. Citation2018c), ca. 7–9 paired dark brown blotches on dorsum (vs. 3–4, N = 5; Bauer Citation2002; Grismer et al. Citation2018c); from C. chrysopylos by DTR ca. 21 (vs. 16–20, N = 17; Bauer Citation2003; Grismer et al. Citation2018c), MVSR ca. 36 (vs. ‘ventral scales’ 37–55, N = 17; Bauer Citation2003; Grismer et al. Citation2018c), nape band absent (vs. present, N = 17; Bauer Citation2003; Grismer et al. Citation2018c), ca. 7–9 paired dark brown blotches on dorsum (vs. 6–8 complete/broken transverse bands on dorsum, N = 17; Bauer Citation2003; Grismer et al. Citation2018c); from C. gansi by PcP 12 (vs. 16–29 in males, N = 4; Bauer Citation2003), ca. 7–9 paired dark brown blotches on dorsum (vs. ca. 10 narrow transverse dark brown bands, N = 6; Bauer Citation2003); from C. jaintiaensis by SVL 65.8 mm (vs. 87.0–96.2 mm, N = 3; Agarwal et al. Citation2018b), SL 10 (vs. 8–9, N = 3; Agarwal et al. Citation2018b), MVSR ca. 36 (vs. 40–42, N = 3; Agarwal et al. Citation2018b), DTR 21 (vs. 19–20, N = 3; Agarwal et al. Citation2018b), ca. 7–9 paired dark brown blotches on dorsum (vs. densely packed connected blotches that form a reticulated pattern over dorsum, N = 3; Agarwal et al. Citation2018b: figs. 9, 12 & 13); from C. montanus by PVT ca. 33 between axilla and groin (vs. 37–43*[15], N = 6; Agarwal et al. Citation2018b; Muansanga et al. Citation2020), PcP 12 (vs. 8–10 in males, N = 3; Agarwal et al. Citation2018b), enlarged tubercles on dorsum of head and eyelids absent [vs. present on holotype and six unvouchered individuals (), not mentioned for paratypes; Agarwal et al. Citation2018b]; from C. myaleiktaung by tubercles absent on dorsum of head (vs. present, slightly enlarged; Grismer et al. Citation2018c), MVSR ca. 36 (vs. 57, N = 1; Grismer et al. Citation2018c), ca. 7–9 paired dark brown blotches on dorsum (vs. six broad transverse cross-bands; Grismer et al. Citation2018c); from C. nagalandensis by PVT ca. 33 between axilla and groin (vs. 34–35, N = 2; Agarwal et al. Citation2018b), DTR ca. 21 (vs. 16–18, N = 2; Agarwal et al. Citation2018b), total TIVLam 17 (vs. 16, N = 2; Agarwal et al. Citation2018b), 12 light bands on tail (vs. 11, N = 1; Agarwal et al. Citation2018b), enlarged tubercles on dorsum of head and eyelids absent (vs. present on holotype, not mentioned for paratype; Agarwal et al. Citation2018b), dorsal tubercles max. four times size of adjacent granular scales (vs. max. five times size of adjacent granular scales on holotype, not mentioned for paratype; assessed from Agarwal et al. Citation2018b: fig. 19c); from C. cayuensis by MVSR ca. 36 (vs. 28–34, N = 19; Li Citation2007), PcP 12 (vs. 6–9, N = 9; Li Citation2007); from C. himalayicus by MVSR ca. 36 (vs. 33–34*[8], N = 2; Agarwal et al. Citation2018a: table 3 & pg. 339; examined material), ventrolateral fold present (vs. absent on holotype, N = 1, presence/absence not noted for referred specimen, ZSIK 19546; Agarwal et al. Citation2018a; examined material), PcP 12 (vs. 10*[9] on male holotype, 10 scales in series with distinct pits but not pores on female referred specimen; Agarwal et al. Citation2018a; examined material); from C. mandalayensis by enlarged tubercles on dorsum of head absent (vs. present, N = 1; Mahony Citation2009; examined material), dorsal tubercles max. four times size of adjacent granular scales (vs. max. five times adjacent granular scale size, N = 1; Mahony Citation2009; examined material), DTR ca. 21 (vs. 18, N = 1; Mahony Citation2009; examined material), MVSR ca. 36 (vs. 32, N = 1; Mahony Citation2009; examined material), PcP 12 in an inverted ‘V’-shaped series (vs. 5 [possibly up to 8] in an inverted ‘V’-shaped series, with an additional enlarged pore-bearing scale situated immediately posterior to the angle of ‘V’-shaped PcP series, N = 1; Mahony Citation2009; examined material); from C. markuscombaii by ca. 7–9 paired dark brown blotches on dorsum (vs. transversely orientated dark brown stripes/elongated blotches, N = 2; Darevsky et al. Citation1998), MVSR ca. 36 (vs. 38–39, N = 2; Darevsky et al. Citation1998), DTR ca. 21 (vs. 14–15, N = 2; Darevsky et al. Citation1998), PcP 12 (vs. 7, N = 1; Darevsky et al. Citation1998); from C. martinstollii by PcP 12 (vs. 0–8, N = 19; Darevsky et al. Citation1998), nuchal band absent (vs. thin and uneven in all specimens in figures, N = 4*[13]; Darevsky et al. Citation1998: ), dorsal blotches dark brown posteriorly, fading anteriorly (vs. uniformly dark brown from anterior to posterior edges on all specimens in figures, N = 4*[13]; Darevsky et al. Citation1998: ); from C. tamaiensis by smaller adult size, SVL 65.8 mm (vs. 90.0 mm, N = 1; Mahony Citation2009; examined material), 12 PcP (vs. 40 PcFP, N = 1; Mahony Citation2009; examined material).
Distribution and natural history
This species is so far only known from the type locality in Namtiram, Manipur (). The specimen was collected during an opportunistic visual survey of herpetofauna. It was located by its eye-shine at ca. 19:00 hours on a tree trunk ca. 1 m above ground level. The tree was in the vicinity of a waterfall surrounded by a moderately dense undergrowth of herbaceous vegetation and three to four small rocky streams flowing into the waterfall. The bordering habitat was primarily secondary forest consisting of bamboo and broad-leaved trees (see also Mahony et al. Citation2020; ). The locality is above a ‘jeepable’ road that connects the towns of Aziuram and Tamenglong in Manipur. RGK has surveyed the locality during three different years but has so far seen and collected only a single specimen indicating that the species might occur in low abundance at the type locality. The type locality is situated at an elevation of 770 m asl. which is just over halfway up the eastern flank of the western-most hill range of the Western Manipur Hills (highest point on this ridge is ca. 1350 m asl., located ca. 35 km south of the collection locality at the southern end of the ridge). Depending on the preferred elevation range of the species, we expect that it may be widespread at least on the eastern flank of the hill range.
Conservation concerns
Obvious threats to the species would be in the form of shifting slash and burn (jhum) farming which is still commonly practiced in the area. Other potential threats could include deforestation for the timber trade. At this point, too little is known about this species’ habitat requirements and geographic distribution to discuss conservation threats in a meaningful way, so officially this species should be categorised as Data Deficient in the IUCN Red List.
Errors in literature
Errors in scientific literature are an unfortunate inevitability, even from the most careful and meticulous authors. The peer review process is intended to act as an additional line of defence against the publication of errors and can often be sufficient to filter out some or many of the obvious issues in manuscripts that may have been overlooked by the authors. The ability of authors to spot their own errors or reviewers to find others’ errors in manuscripts varies widely depending on the individuals’ personal abilities, their experience, frame of mind at the time of carrying out the work (writing or reviewing), and dedication and/or interest to ensure a high level of accuracy in the work. Often errors can occur due to misunderstandings or miscommunications between co-authors as to whom is responsible for cross-checking data presented in the manuscripts before final submission, or when changes are made to basic data in one part of a manuscript, but not corrected elsewhere (e.g. between text and tables). Errors such as typos can occur when copying data from referred literature, or between different sections of the same paper, when reformatting tables, and from glitches in software. Other less obvious errors can occur through the misinterpretation of data between studies and copying published errors from other referred literature. Spotting errors in the text of manuscripts can be relatively easy sometimes (e.g. misidentifications of species), however, spotting errors that involve numbers, or ranges of numbers, can be a lot more challenging as it necessitates the extremely tedious task of carefully cross-checking the data from multiple sources. Reviewers cannot be expected to extensively cross-reference character details either within manuscripts or between a reviewed manuscript and cited reference, so the responsibility lies with the authors to ensure their data is accurate. This may seem very obvious and a standard necessity in scientific writing, but unfortunately due diligence by authors is often insufficient, or even neglected.
During the preparation of the morphological comparisons, distribution map and molecular phylogenetic sections of this paper, a worrying number of errors were found in the literature involving species of the gansi group and incertae sedis species in the Indo-Burma radiation. Some errors are minor and perhaps trivial (e.g. abbreviations for morphological characters differing between materials and methods sections and the text/tables) but there are errors (e.g. involving morphological characters and measurements) that simply cannot be ignored, and these errors have already been reproduced in subsequent publications jeopardising the accuracy of future taxonomic research on this group. It is absolutely not our intention to name and shame authors (SM has authored/co-authored some of the papers), and readers should certainly not imagine that the kinds of errors identified here are unique to papers that only deal with this particular taxonomic group. These types of errors are unfortunately rampant in scientific literature, and this section should serve as a call for better vigilance in manuscript preparation and peer review for all who work in our field. The following are only the errors that were observed casually (complete critical review is beyond the scope of this paper) and should not be considered exhaustive even for these papers: Agarwal et al. (Citation2018a, Citation2018b); Annandale (Citation1906); Darevsky et al. (Citation1998); Grismer et al. (Citation2018a, Citation2018b, Citation2018c, Citation2018d, Citation2019a, Citation2019b); Mahony (Citation2009); Muansanga et al. (Citation2020); Purkayastha et al. (Citation2020); Wood Jr et al. (Citation2012). The errors, corrections, and the justifications thereof, are provided below, arranged by species:
1) Cyrtodactylus aunglini MVSR: Grismer et al. (Citation2018c) gave the ‘ventral scales’ counts as ‘41–49’ in the ‘Diagnosis’ section of the species description (pg. 158), as ‘47–49’ in table 3 (pg. 160) and as ‘41–47’, N = 10 in table 4 (pg. 162), the ‘Comparison’ section (pg. 164) and table 5 (pg. 165). The holotype was stated to have 41 ‘ventral scales’ (pg. 159), and table 5 (pg. 165) gave the per specimen meristic counts, thus we consider the correct range for the species for ‘ventral scales’ as 41–47, N = 10. The comparison table given for C. mombergi Grismer, Wood, Quah, Thura, Herr and Lin, 2019 in the original description (Grismer et al. Citation2019b: table 2, pg. 8 and ‘Comparison’ section, pg. 22) gave the erroneous range of ‘ventral scales’ as ‘47–49’ for C. aunglini and is highlighted as a diagnostic character. Despite this error, C. aunglini and C. mombergi do not have overlapping ventral-scale ranges when correcting the range for C. aunglini (41–47 vs. 31–39). It is not clear why Grismer et al. (Citation2019b: table 3, pg. 8–10) gave ‘N = 6’ for the number of specimens of C. aunglini when counts given in Grismer et al. (Citation2018c) represented 10 specimens.
2) C. aunglini PVT: Grismer et al. (Citation2018c) gave the PVT ‘36–45’, N = 10 in two tables (tables 3 & 4) and in the ‘Diagnosis’ section (pg. 158), however, Grismer et al. (Citation2018c: table 5) gave the meristic counts for each specimen in the type series where the range is clearly PVT 37–45, N = 10, thus we consider PVT 37–45 to be the correct range for the species.
3) C. aunglini type locality: Grismer et al. (Citation2018c: pg. 158) gave the type locality for the species as ‘Kyauk Nagar Cave, 11 km southwest of Pyin Oo Lwin, Pyin Oo Lwin Township, Pyin Oo Lwin District, Mandalay Region, Myanmar (20.93087°N, 95.22580°E; 715 m in elevation)’. The same coordinates were given for the type locality in the caption of and 3. When preparing our distribution map (), the GPS coordinates (20.93087°N, 95.22580°E) placed the type locality in the vicinity of Mt. Popa National Park, Nyaung-U District, Mandalay Region (the type locality of C. brevidactylus), ca. 170 km southwest of the Pyin Oo Lwin location, so it appears that the authors might have mixed up the coordinates for these two species. We have estimated the coordinates on Google Maps for the purpose of preparing our distribution map (, Appendix II) pending clarification by the original authors.
4) C. brevidactylus post cloacal spurs: Bauer’s (Citation2002: pg. 77) original description for the species stated ‘cloacal spurs 3–4’. Grismer et al. (Citation2018c: table 3, pg. 160, & Citation2019b: table 2, pg. 9) both stated that data for C. brevidactylus was derived from Bauer (Citation2002) supplemented with data from two additional specimens (LSUHC 13498–99) but stated ‘4–7’ postcloacal spurs for the species. No explanation was given for the differing counts provided in Grismer et al. (Citation2018c & Citation2019b; no individual counts were provided for the two LSUHC specimens), thus we regard the range of 4–7 postcloacal spurs as erroneous pending further clarification.
5) C. gansi SVL: Bauer (Citation2003: table 3, pg. 478) gave the max. SVL ‘62.4’ mm, N = 6. Grismer et al. (Citation2018c: table 3, pg. 160, & Citation2019b: table 2, pg. 8) stated that data were taken from Bauer (Citation2003) only, but erroneously provided a max. SVL of ‘62.3’ mm, and stated the range was based on seven specimens (‘N = 7’—see point 6 below). Grismer et al. (Citation2018c: pg. 164) again gave a max. SVL of ‘62.3’ mm in the ‘Comparisons’ section of C. aunglini.
6) C. gansi specimen numbers: Bauer (Citation2003: pg. 475) listed the following five paratypes: CAS 222411, USNM 559839, CAS 226145, CAS 222412–222413, however, in table 3 and in the ‘Variation’ section (pg. 478) he provided measurements and discussions for five paratypes excluding USNM 559839, but including ‘CAS 226144’. According to the CAS collection database (http://researcharchive.calacademy.org/research/herpetology/catalog/index.asp), CAS 226144 (field number JBS 8258) was exchanged to the USNM where it was given the specimen number USNM 559839. The USNM (now NMNH) collection database (https://collections.nmnh.si.edu/search/herps/) lists this specimen as present in the collection with the number NMNH 559839 (field number JBS 8258). The type series of C. gansi is therefore comprised of six specimens and the character summary in Grismer et al. (Citation2018c: table 3) should read N = 6 (not ‘N = 7’).
7) C. gansi MVSR: Bauer (Citation2003: pgs. 477 & 478) stated MVSR as ‘36–40’, however, Grismer et al. (Citation2018c: table 3, pg. 160, & Citation2019b: table 2, pg. 8) gave the range as ‘30–36’. This error is repeated in the comparison section of C. aunglini (Grismer et al. Citation2018c: pg. 164). Despite the error, C. aunglini and C. gansi do not have overlapping ventral-scale ranges when correcting the range for C. gansi (41–47 vs. 36–40).
8) C. himalayicus MVSR: Agarwal et al. (Citation2018a: table 2) gave the MVSR range as ‘33–44’, but clearly stated ‘33 or 34’ in the ‘Diagnosis’ (pg. 339) of C. himalayicus and in their table 3 as 33 and 34 for the MVSR values of the two examined specimens. We have examined the two specimens and can confirm the correct range is 33–34 MVSR. Agarwal et al. (Citation2018b: table 3) gave the erroneous range of MVSR ‘33–44’ in their diagnostic character summary table presumably based on the error in Agarwal et al. (Citation2018a: table 2).
9) C. himalayicus PcP: Agarwal et al. (Citation2018a: table 3) stated PcP as 8 for the holotype, but in table 2 and the species ‘Diagnosis’ (pg. 340) they stated PcP as 10. Annandale (Citation1906) stated the holotype had PcP 11. We reconfirm here based on direct observation of the holotype that PcP 10 is correct for this specimen.
10) C. jaintiaensis figure captions: The images for figures 12 and 13 in Agarwal et al. (Citation2018b) were accidentally switched so do not match their captions, i.e. the specimen in Agarwal et al. (Citation2018b: fig. 12) is not the male paratype, BNHS 2246 but that of the female holotype, and the specimen in Agarwal et al. (Citation2018b: fig. 13) represents the paratype BNHS 2246 and not the holotype as incorrectly stated in the figure captions.
11) C. mandalayensis type locality: Mahony (Citation2009: pgs. 68 & 69) erroneously gave the GPS coordinates for the type locality of C. mandalayensis as ‘(17°34ʹ0’N, 95°5ʹ0″Eʹ), which refers to Moe goke, a small village tract in the Lemyethna Township, Hinthada District, Ayeyarwady Region of southwestern Myanmar. The collection locality of the holotype, ‘Magok, on the Irawadi, northeast of Mandalay, Upper Burma’, refers to Mogok (but also spelt ‘Moegoke’), a town that lies ca. 613 km to the northeast of the coordinates given by Mahony (Citation2009). This is a good example for why GPS coordinates obtained for place names should always be plotted to ensure they apply to the correct locality, which Mahony (Citation2009) clearly did not do (SM apologises for any confusion caused by this error). The type locality also contains information that is both misleading (‘on the River Ayeyarwadi’—Mogok is located ca. 54 km east of the Ayeyarwady River) and unnecessary (‘north-east of Mandalay’), so we here provide the corrected and refined type locality of Cyrtodactylus mandalayensis as, ‘Mogok Town (ca. 22.922896, 96.506150, ca. 1170 m asl.) or its vicinity, Mogok Township, Pyin Oo Lwin District, Mandalay Region, northern Myanmar’.
12) C. martinstollii SVL: Darevsky et al. Citation1998: pg. 89) gave the SVL in the species ‘Diagnosis’ as a maximum of 82 mm, and the holotype is stated to have SVL 82 mm in the species description (pg. 89), but in the summary table of characters (table 2, pg. 93), the max. SVL is given as 78 mm. The error in table 2 may have been derived from the max. SVL given for the male paratype series in table 1 (pg. 91), despite this table stating the max. SVL of the female paratype series as 79 mm. Therefore, SVL 82 mm is considered to be the correct max. SVL for the species.
13) C. martinstollii figure caption, specimen number and correct spelling: The caption to in Darevsky et al. (Citation1998) stated that the ‘Second specimen from the left is the holotype’. The caption of claimed that the specimen on the left is also the holotype; however, dorsal markings for the specimen in distinctly match the specimen on the far left of (this specimen is also the same animal in the image in life, fig. 8). A photograph of the holotype (MHNG 2590.009: number given incorrectly as ‘MHNG 2590.09’ in Darevsky et al. Citation1998) was recently provided to SM (Schmitz pers. comm. 2020) allowing us to confirm that the caption for in Darevsky et al. (Citation1998) correctly identified the holotype, thus the specimen in their and 8 represent a paratype (current specimen number not ascertained). Note that the original spelling for the species name is C. martinstollii not C. ‘martinstolii’ or C. ‘martinstolli’ as erroneously referred in many studies (e.g. Mahony Citation2009; Mahony et al. Citation2009b; Bauer et al. Citation2013; Agarwal et al. Citation2018a, Citation2018b, Citation2018c; Barabanov and Doronin Citation2020; Purkayastha et al. Citation2020). The incorrect spelling C. ‘martinstolli’ is at the time of writing given for the species on The Reptile Database (Uetz Citation2021 online, accessed on 11 December 2020; hereafter TRD). TRD is the go-to database of virtually all who work on reptiles, but due to the enormously ambitious scale of such a project, extensive erroneous information exists within the pages, e.g. on the species account page for C. martinstollii, the chresonymy includes six examples of alternative binomial combinations, including two spellings for the species epithet from literature, but when cross-checked against the cited literature, all examples on the webpage contain at least one error. The extent of errors in this example is hopefully an exception on TRD (an otherwise indispensable database resource), but emphasises that databases such as this must not be relied upon in lieu of referring to the original literature cited in the database. Despite this, databases such as TRD are increasingly being cited instead of peer reviewed original literature in scientific papers, a practice that should be discouraged by journal editors and reviewers to reduce the propagation of errors in literature.
14) C. montanus MVSR: Agarwal et al. (Citation2018b) correctly provided the MVSR range of 36–42 in tables 3 and 4, but erroneously gave ‘21–23’ MVSR in the species ‘Diagnosis’ (pg. 524). Grismer et al. (Citation2018c: table 3) used the erroneous range of ‘21–23’ MVSR for the species and as a result they incorrectly stated ‘21–57 ventral scales’ in their definition of the gansi group, which should have read ‘32–57 ventral scales’ because C. mandalayensis has 32 MVSR (N = 1, Mahony Citation2009). Grismer et al. (Citation2019b: pg. 6) again gave ‘21–57 ventral scales’ for the definition of their expanded ‘gansi group’ (equivalent to the khasiensis clade as proposed in this study) despite correcting the ventral scale count for C. montanus to ‘36–42’ in their table 2. The correct MVSR range for their expanded ‘gansi group’ (equivalent to our khasiensis clade) should be 31–57 based on the lower limit provided for C. mombergi (‘31–39 ventral scales’, N = 13; Grismer et al. Citation2019b).
15) C. montanus meristics: For their referred specimen, Muansanga et al. (Citation2020) stated ‘39 longitudinal rows of rounded dorsal tubercles, 23 paravertebral tubercles, 23 ventral scales between ventrolateral folds’. These counts are outside the limits of what is known for C. montanus (DTR 21–23, PVT 37–43 and MVSR 36–42; Agarwal et al. Citation2018b). It appears that Muansanga et al. (Citation2020) mixed up their DTR and PVT numbers. Their MVSR falls within the limits of the error in Agarwal et al. (Citation2018b, pg. 524) discussed above in point 14. The authors neither explained their methods for meristic counts, nor did they refer to the species’ original description (Agarwal et al. Citation2018b) in their text, so it cannot be determined how they obtained the provided count, or if they read the original description at all.
16) C. montanus voucher locality: Muansanga et al. (Citation2020: ) provided a map depicting the range extension of their new record against the type locality. The type locality of C. montanus was erroneously depicted ca. 20 km north of the actual type locality, and the locality of the new specimen was depicted ca. 10 km southwest of the provided coordinates. Although such discrepancies in localities may appear small, the topography of the region is comprised of north-south orientated mountain ridges separated by deep valleys and the mapped point for the new record is located on a different mountain side. The authors clearly measured the distance between the GPS coordinates of the two localities (given as 20.1 km) but their map, assuming their scale bar is correct, shows the two localities as > 40 km apart. The importance of accurately mapping GPS coordinates cannot be overstated, particularly for Cyrtodactylus species that are often found to be microendemics.
17) C. nagalandensis MVSR: Agarwal et al. (Citation2018b) stated correctly 36–37 MVSR in tables 3 and 4 and in the ‘Comparisons’ section (pg. 531), but erroneously gave as ‘34 or 35’ MVSR in the species ‘Diagnosis’ (pg. 528).
18) C. pyinyaungensis Grismer, Wood, Thura, Zin, Quah, Murdoch, Grismer, Lin, Kyaw and Lwin, 2018, specimen number: Grismer et al. (Citation2018a: pg. 877) provided the same specimen number ‘LSUHC 13149’ for both the holotype and one paratype in their description of C. pyinyaungensis. Elsewhere (e.g. pgs. 879–880, 881), the authors referred to a specimen as paratype (LSUHC 13148) that was not listed as a type on pg. 877. The holotype was also referred to elsewhere as ‘LSUHC 13149’ (e.g. pgs. 878–880). It is apparent that there was an error on pg. 887 where the paratype number ‘LSUHC 13149’, should have read ‘LSUHC 13148’. Also, the paratype ‘BYU 52234’ listed on pg. 877 was stated to have the alternative specimen number ‘LSUHC 12939’ in the caption for figure S1 (pg. 958). This important information should have been presented in the list of paratypes (on pg. 877) as it is the source of further confusion with regards to sequence-related data (see point 20 below). Museum catalogue numbers for type specimens should always be accompanied by other associated specimen numbers (e.g. field tag numbers) at least once in the original description of a species.
19) C. aunglini, C. chrysopylos and C. myaleiktaung GenBank/sequence data errors: Grismer et al. (Citation2018c: ) provided the same GenBank number ‘MH764589’ for two specimens, C. aunglini (LSUHC 13948) and C. myaleiktaung (LSUHC 13965). On GenBank, the sequence identification number MH764589.1 corresponds with C. aunglini (LSUHC 13948) and the details given for the sequence identification number MH764598.1 refer to C. myaleiktaung (LSUHC 13965). However, the latter sequence is 99–100% identical to other sequences referred to C. chrysopylos () submitted by Grismer et al. (Citation2018c; i.e. MH764600.1, MH764603.1); ‘MH764598’ was assigned to C. chrysopylos (LSUHC 13126) in Grismer et al. (Citation2018c: ). At the time of writing (16 September 2020), there is no sequence on GenBank referable to C. myaleiktaung. Despite this, Purkayastha et al. (Citation2020: ) included C. myaleiktaung in their phylogeny, listing the GenBank number ‘MH764598’; however, their phylogenetic tree () indicated that the sequence was deeply diverged from the sequence they included for C. chrysopylos (JX440531—BLAST of the two sequences shows they are 99.7% similar). Purkayastha et al. (Citation2020) must have identified the issue highlighted above and obtained the correct sequence from elsewhere for C. myaleiktaung but did not provide the correct information in their table or elsewhere in their paper for the source of the sequence they used for this species. Purkayastha et al. (Citation2020) did not cite Grismer et al. (Citation2018c) as one of the listed references for sequence data and did not mention any issues related to this sequence. On GenBank, the identification number MH764595.1 is assigned to C. chrysopylos (LSUHC 13126). On GenBank and in Grismer et al. (Citation2018c), the details given for the sequence identification number MH764590.1 is C. chrysopylos (LSUHC 13938); however, this sequence is 99–100% identical to other sequences referred to C. aunglini on GenBank (e.g. MH764589.1, MH764603.1). What is clear from these examples is that there had been a complicated mislabelling of sequences generated in Grismer et al. (Citation2018c) both on GenBank and in the paper itself. Through correspondence with the original authors, we have been informed that they have now corrected the relevant records on GenBank (Grismer pers. comm. 21 March 2021).
20) C. pyinyaungensis GenBank/sequence data errors: Grismer et al. (Citation2018a: , pg. 867) generated one sequence for C. pyinyaungensis from a paratype, BYU 52234 (= LSUHC 12939) with the GenBank number MF872307. Grismer et al. (Citation2018d: ) generated sequences for three specimens (LSUHC 13139, LSUHC 13148, LSUHC 13149) of C. pyinyaungensis with the GenBank numbers MH624119, MH624120 and MH624121 (respectively), however, on the phylogenetic tree (Grismer et al. Citation2018d: ), the specimen number ‘LSUHC 13139’ was inexplicably replaced with ‘LSUHC 12939’ (= BYU 52234). Grismer et al. (Citation2019a: ) included three sequences of C. pyinyaungensis in their phylogenetic tree, labelled ‘LSUHC 13148 (MH624120)’ and ‘LSUHC 13149 (MH624121)’ as given earlier in Grismer et al. (Citation2018d), and ‘LSUHC 12939 (MH624119)’, for which the specimen number ‘LSUHC 13139’ was given in Grismer et al. (Citation2018d: ). The specimen number ‘LSUHC 13139’ appears nowhere else in Grismer et al. (Citation2018a; Citation2018d or Citation2019a) and so we presume this to be an error that should have read ‘LSUHC 12939’ in Grismer et al. (Citation2018d: ). If this is correct, the same paratype specimen LSUHC 12939 (= BYU 52234) was sequenced twice and given the GenBank number MF872307 in Grismer et al. (Citation2018a) and MH624119 in Grismer et al. (Citation2018d). On the NCBI database (accessed on 17 November 2020), when searching for the four GenBank numbers, MF872307, MH624119, MH624120 and MH624121, the first number corresponds with details given in Grismer et al. (Citation2018a: ), as it should; however, the numbers MH624119–MH624121 given in Grismer et al. (Citation2018d & Citation2019a) apply to the bacteria ‘Lactobacillus plantarum’. According to the NCBI database (accessed on 17 November 2020), the correct GenBank numbers for the sequences generated in Grismer et al. (Citation2018d, and incorrectly cited in Grismer et al. Citation2019a) are: LSUHC 12939 (MH756187.1), LSUHC 13148 (MH756189.1) and LSUHC 13149 (MH756188.1).
21) C. feae (Boulenger, Citation1893), C. myintkyawthurai Grismer, Wood, Quah, Murdoch, Grismer, Herr, Espinoza, Brown and Lin, 2018, and C. peguensis (Boulenger, Citation1893), GenBank/sequence data errors: GenBank number JX440536 (Wood Jr et al. Citation2012) represents specimen number USNM 559805 and was identified in the original paper and on GenBank as Cyrtodactylus feae. This sequence was included in the phylogeny of Grismer et al. (Citation2018a) and stated to have been misidentified in Wood Jr et al. (Citation2012) and Agarwal et al. (Citation2014) with the following comment ‘[the sequence] … was reported to L.L.G. to be C. peguensis (G. R. Zug, unpubl. data)’. In addition to the aforementioned studies, the sequence (JX440536) had been used in phylogenetic analyses in several other papers under the identity of C. feae (e.g. Harvey et al. Citation2015, Citation2016; Brennan et al. Citation2017). This sequence was subsequently included in the phylogeny of Grismer et al. (Citation2018d: ) where it was found to represent C. myintkyawthurai; however, Grismer et al. (Citation2018d) made no indication in the text to clarify the identity of this sequence or their earlier comment in Grismer et al. (Citation2018a). Most recently, Purkayastha et al. (Citation2020) again referred to this sequence incorrectly as C. feae in their analyses, demonstrating the importance of clearly correcting misidentified sequences when discovered to help prevent subsequent errors in literature.
22) C. ayeyarwadyensis Bauer, Citation2003, GenBank/sequence/museum database errors: Wood Jr et al. (Citation2012: ) provided the specimen number ‘CAS 216459’ for a C. ayeyarwadyensis specimen associated with the following GenBank numbers: JX440526.1 (ND2), JX440581.1 (MXRA5), JX440634.1 (PDC), JX440685.1 (RAG1); the same specimen number is associated with the GenBank record on the NCBI database (accessed on 17 November 2020). This specimen number has also been associated with these sequences in subsequent studies (e.g. Agarwal et al. Citation2014, Citation2018a, Citation2018b; Brennan et al. Citation2017). According to the CAS herpetology collection database (http://researcharchive.calacademy.org/research/herpetology/catalog/index.asp; accessed on 17 November 2020), the specimen number CAS 216459 was associated with a specimen of ‘Micrixalus [Ingerana] borealis’ (Annandale Citation1912). Wood Jr et al. (Citation2012: ) provided the full locality details of the sequenced specimen as ‘Myanmar, Rakhine State, Than Dawe District, Gwa Township, Rakhine Yoma Elephant Range, Elephant Camp’. A search of the CAS collection database for tissue sampled specimens of C. ayeyarwadyensis produced a list of 10 specimens, one of which is from the collection locality given in Wood Jr et al. (Citation2012), CAS 216506, which might at first be thought to be the correct number for the sequenced sample. The authors of Wood Jr et al. (Citation2012) have subsequently confirmed that the specimen sequenced was neither CAS 216459 nor CAS 216506, but CAS 212459 from ‘Myanmar, Ayeyarwady Division, vicinity of Mwe Hauk village 16º16ʹ29.4“ N, 94º46ʹ04.0” E’, the original confusion arising from a simple accidental change of the second ‘2’ in the specimen number to a ‘6’ in a lab book, a very easily made error (Bauer, Jackman & Wood, pers. comm. 24 November 2020). Compounding the confusion was that this specimen (CAS 212459) is not listed on the CAS collection database as possessing a tissue sample, and so was not included in the aforementioned list of ten specimens; however, the correct GenBank numbers are listed in the CAS database for this specimen under the identifier ‘GenBank#’, implying an omission on the CAS collection database for this specimen under the ‘Tissue’ identifier (CAS Collections Manager has been notified).
The kinds of errors highlighted here are very common on GenBank, in publications, and in online museum collection databases. They can be very difficult to identify and are definitely not restricted to the examples provided in points 19–22 (e.g. see similar errors highlighted for seven papers in Mahony et al. Citation2017: supplementary information, table S3). In addition to the obvious compounding of errors in subsequent publications, errors such as these could result in the unnecessary duplication of sequencing efforts in the future for the same tissue sample, diminishing an already very limited and important analytical resource. To help avoid the duplication of errors such as these in future analyses(/publications), we recommend that authors, reviewers and editors check the following details on manuscripts prior to publication: A) GenBank sequence identification numbers should be correctly cited by including the version number given as a decimal point (e.g. underlined in this example: MH764590.1). This ensures that the most up-to-date version of the sequence is being used in analyses (Clark et al. Citation2016; Benson et al. Citation2017); B) Data associated with GenBank sequences should be cross-checked against the publications that generated them to make sure that the information matches. GenBank sequence identification numbers referred to in all publications should cite the original publication from which the sequence was generated. NCBI specifically states ‘When citing data in GenBank, it is appropriate to give the sequence name, primary accession number, and the publication in which the sequence first appeared’ (https://www.ncbi.nlm.nih.gov/genbank/release/current/, accessed 12 December 2020), a basic request in the terms of use set out by GenBank that has been ignored by most users of the database. Citing GenBank only should not be considered acceptable since GenBank did not generate any of the sequences. Not providing a citation and reference for original data used in analyses is technically and scientifically unethical, and directly damages the field of study by undermining the importance of papers that produce original data (through direct loss of citations); C) The topology of newly generated phylogenies should be compared to similar previously published trees to check that sequences downloaded from GenBank represent what they are supposed to. A subsequent BLAST search of questionable sequences against the GenBank repository can help to identify what type of error/s occurred; D) Any errors discovered should be notified in publication so that subsequent studies do not compound the problem by duplicating errors or cause subsequent researchers to spend inordinate amounts of their time trying to figure out a tangled mess of errors in others’ publications; E) The original authors of errors involving GenBank records should be notified and requested to make necessary corrections to their GenBank records. Currently, GenBank only permits original authors of sequences to update/correct errors on records, so authors of sequences accessioned on GenBank are therefore obliged to correct errors associated with their sequences. This includes updating species identifications on GenBank as new species are described for which sequences are available.
23) Morphological character summary table errors: Grismer et al. (Citation2018c: table 3) provided a summary table of morphological characters for their ‘gansi group’ but did not cite the source for data given in their table for Cyrtodactylus jaintiaensis, C. montanus and C. nagalandensis. Presumably, this data came from Agarwal (in lit.) since Agarwal et al. (Citation2018b) was published only a couple of weeks before Grismer et al. (Citation2018c), perhaps preventing the latter study sufficient time to update information in their summary table. Numerous characters given in Grismer et al. (Citation2018c: table 3) for these three species (e.g. PVT, total subdigital lamellae on 4th toe, PcP, etc.) differed considerably from the data given in Agarwal et al. (Citation2018b) and should be considered erroneous. Subsequently, Grismer et al. (Citation2019b: table 2) provided meristic ranges that corresponded with some of those provided in Agarwal et al. (Citation2018b); however, several errors were repeated (e.g. PcP of C. nagalandensis). The different methodology on how scale counts were made between these studies also means that the meristic ranges should not have been considered directly comparable, as the following examples demonstrate: A) total lamellae on toe IV in Agarwal et al. (Citation2018b) excluded small scales between the basal and apical lamellae series, whereas, total lamellae on toe IV counts in Grismer et al. (Citation2018c, Citation2019b) included these small scales (‘ … including the granular scales adjacent to the digital inflection’); B) supralabial counts given in the comparison tables in Grismer et al. (Citation2018c, Citation2019b) for the species they examined were counted to the midorbital position, but they compared their counts directly with the total supralabial counts for taxa described by Agarwal et al. (Citation2018a & Citation2018b), despite the latter authors also providing counts to the midorbital position (Agarwal et al. Citation2018a: table 3 & Citation2018b: table 4); C) Grismer et al. (Citation2018c, Citation2019b) directly compared PVT counts between their examined specimens and those given in Bauer (Citation2002, Citation2003) and Agarwal et al. (Citation2018a) despite these counts having been made differently, i.e. Bauer (Citation2002, Citation2003) and Agarwal et al. (Citation2018a) counted PVT from ‘occiput to mid sacrum’, whereas Grismer et al. (Citation2018c, Citation2019b) counted ‘the number of paravertebral tubercles (PVT) between limb insertions counted in a straight line immediately left of the vertebral column’. Agarwal et al. (Citation2018a: pg. 335) had clearly stated in the ‘Materials and methods’ section that PVT counts are not comparable with the methodology used in Grismer et al. papers. Overlooking considerable differences in methods when comparing meristic and/or mensural data between studies results in unusable character summary tables and erroneous comparison sections. The sources of data provided in summary tables also requires careful scrutiny, e.g. Grismer et al. (Citation2019b: table 3) cited Mahony et al. (Citation2009b) for morphological data given for C. ayeyarwadyensis, despite Agarwal et al. (Citation2018a: pg. 353) referring to the specimens described by Mahony et al. (Citation2009b) as needing further taxonomic study in light of their geographic proximity to the morphologically similar C. tripuraensis Agarwal, Mahony, Giri, Chaitanya and Bauer, 2018. For all of the above reasons (and other errors not discussed in detail, e.g. PcP of C. mandalayensis erroneously given as ‘8’, etc.), we recommend not using character summary tables published in Grismer et al. (Citation2018c, Citation2019b) as a source for meristic comparisons in future studies. We have been recently informed that the authors have a manuscript in preparation rectifying these errors (Grismer pers. comm. 24 March 2021).
The above examples demonstrate that many of the errors in meristics and morphometric data appear when the same data is presented in multiple sections of the same paper, particularly in character summary tables and species ‘diagnosis’ sections. These examples therefore demonstrate the criticality of reading taxonomic papers thoroughly and not just scanning through. Morphological character summary tables are extremely popular in taxonomic papers, sometimes even insisted upon by reviewers and/or editors. The intention of giving such tables is to provide the reader with a single easy source for all of the relevant morphological characters of related species, typically combining data from multiple sources. If these tables are made with a lot of care, they are certainly useful, but should never be considered a one-stop source as more often than not, careful scrutiny of these tables will demonstrate that they contain errors. Unfortunately, the apparent convenience of these tables encourages many taxonomists to rely solely upon summary tables rather than putting in the expected work of referring directly to the original sources of the data when identifying newly collected specimens, or when diagnosing new species. To reduce the propagation of errors in taxonomic literature, we make the following suggestions for consideration by reviewers and editors involving the publication of morphological character summary tables: 1) not permit authors to cite summary tables as the sole source of morphological character data, thus compelling researchers to refer to the original descriptions or other relevant literature that present original data. Likewise, readers should only use character summary tables as a guide to inform them of which original papers they need to both cite and carefully cross-check data from; 2) not encourage or insist authors who do not have extensive demonstrated knowledge of both the species and relevant literature on a taxonomic group to include a morphological character summary table in their manuscript; 3) when editors and reviewers are presented with a character summary table in a manuscript, reviewers may be asked to demonstrate that they have cross-checked for errors a reasonable random portion of characters from the table against the original literature.
Cyrtodactylus in Manipur State, northeast India
Until recently, northeast India had remained poorly studied with regard to the taxonomy and systematics of its native herpetofauna. Recent dedicated surveying efforts coupled with integrative taxonomic studies have begun to shed light on its importance as a region of high taxonomic endemicity (e.g. Kamei et al. Citation2013; Biju et al. Citation2016; Mahony et al. Citation2018, Citation2020; Agarwal et al. Citation2018a, Citation2018b; Giri et al. Citation2019a, Citation2019b; Purkayastha et al. Citation2020). Historically, the herpetofauna of the region encompassing the present Manipur State did not receive as much attention as other northeast Indian states (Acharji and Kripalani Citation1951; Singh Citation1977; Murthy and Singh Citation1982). In modern times also, Manipur’s herpetofauna has remained poorly studied with only a handful of publications based on recent field research (e.g. Mathew Citation2005; Sarkar et al. Citation2005; Ningobam and Bordoloi Citation2007; Kamei et al. Citation2009; Mathew and Sen Citation2009; Biju et al. Citation2016; Garg et al. Citation2018; Mahony et al. Citation2018, Citation2020). Reasons for the lack of herpetofaunal surveys in Manipur may be varied and complex but regular ethno-civil strife, insurgency and militancy have impeded work (Kamei Citation2017), particularly for night-time surveys in the countryside.
To our knowledge, this study is the first to include molecular data for a gecko from Manipur State, and Cyrtodactylus namtiram sp. nov. currently represents the only endemic species of reptile in the state. Mathew (Citation2005) reported one specimen (NERC/ZSI museum catalogue number not provided) of Cyrtodactylus ‘khasiensis’ from ‘Litan’ Village (ca. 24.9417, 94.2131; GPS coordinates estimated from Google Maps), Ukhrul District, in eastern Manipur, collected by H. Khajuria on 20 January 1977. Rather than providing a description of this specimen, Mathew (Citation2005) unfortunately only gave a brief morphological summary of the composite species C. ‘khasiensis’ sensu lato, plagiarised directly from Smith (Citation1935). We have not examined this specimen but consider it very unlikely to represent C. khasiensis sensu stricto as this species is now considered to be endemic to the southern Khasi Hills (Agarwal et al. Citation2014, Citation2018a, Citation2018b). We encourage that the Ukhrul specimen be examined to determine its taxonomic identity, though it should be clear that reliable species-level identifications of most northeast Indian Cyrtodactylus populations may only be attainable with the aid of molecular techniques such as DNA barcoding to prevent future misidentifications. Considering that most of the Cyrtodactylus species identified in northeast India are known to be endemic to small geographic areas (e.g. Al-Razi et al. Citation2018; Agarwal et al. Citation2018a, Citation2018b; Muansanga et al. Citation2020), providing range extensions for these species should be done with great care to prevent misidentifications which would have negative impacts for determining the conservation needs of potentially threatened species.
Addendum. During the second review of this paper, five additional new species of Cyrtodactylus from the khasiensis clade were described from northeast India (Kamei and Mahony Citation2021; Purkayastha et al. Citation2021). Three of these are members of the khasiensis group from eastern Meghalaya State, and the other two are members of the gansi group from Mizoram State. To demonstrate that these descriptions do not affect the validity of the new species described herein, we here provide a morphological comparison with the two new gansi group species. Cyrtodactylus namtiram sp. nov. differs from C. aaronbaueri Purkayastha, Lalremsanga, Bohra, Biakzuala, Decemson, Muansanga, Vabeiryureilai, Chauhan & Rathee, Citation2021, which is represented in our molecular phylogeny as ‘C. sp. Mizoram’ in , by PcP 12 (vs. 7–8, N = 6), DTR ca. 21 (vs. 22–28, N = 9), and PVT ca. 33 between level of axilla and level of groin (vs. 36–39, N = 9); and from C. bengkhuaiai Purkayastha, Lalremsanga, Bohra, Biakzuala, Decemson, Muansanga, Vabeiryureilai, Chauhan & Rathee, Citation2021 (not represented in our phylogenetic analyses) by PcP 12 (vs. 5–7, N = 3), DTR ca. 21 (vs. 22–26, N = 6), PVT ca. 33 between level of axilla and level of groin (vs. 35–41, N = 6), and MVSR ca. 36 (vs. 37–42, N = 6). A BLASTn search against GenBank demonstrates that the ND2 sequence for Cyrtodactylus namtiram sp. nov. is most similar to C. bengkhuaiai (8.9–12.7% different) which is similar to the differences observed between closely related taxa in the gansi group (e.g. 10.3–11.3% between C. aaronbaueri and C. montanus).
Acknowledgements
RGK thanks the Forest Department, Government of Manipur for fieldwork and specimen collection permit (No. 3/22/2006-WL/87 dated 27 May 2013 & No. 3/22/2006-WL/9 dated 7 April 2014). This research was supported by a Rufford Foundation Small Grant (15255-1) to RGK, and Irish Research Council-Marie Skłodowska-Curie CAROLINE Fellowship (CLNE/2017/482) to SM. For access to molecular lab facilities and lab support, RGK thanks Praveen Karanth (Indian Institute of Science, Bangalore, India) and his students Chinta Sidhartha, Kunal Arekar, and Maitreya Sil. RGK thanks L.L. Aaron Riamei, Keibianliu Riamei and Lungchung Riamei for hosting her in their home during fieldwork, Pourangteu Riamei for assistance in the field, and Janghemlung Panmei for providing a jeep and driver for her July 2015 field expedition. Thanks to Lee Grismer for providing the correct sequence for C. myaleiktaung used in our molecular analyses, Perry Lee Wood, Jr., Todd Jackman and Aaron Bauer for clarification of the C ayeyarwadyensis sequence error, and Aaron Bauer for confirming the spelling of C. martinstollii in a paper (Rösler, H. 2000. Gekkota, 2: 28–153), which was unavailable to us. For granting access to collections and curatorial assistance, SM thanks Colin McCarthy (NHMUK), G. Ramakrishna and B.H.C.K. Murthy (ZSIK), as well as Andreas Schmitz (MHNG) for providing images of the C. martinstollii holotype. RGK thanks Rahul Khot for accessioning the type material in the BNHS museum. Thanks also to Peter Uetz and The Reptile Database team (https://reptile-database.org/) for the exceptional contribution to the herpetological community that they provide. We thank the four anonymous reviewers and the editor for their valuable critical review of this paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Acharji MN, Kripalani MB. 1951. Contributions to the fauna of the Manipur state, Assam. Part IV. Reptilia. Rec Indian Mus. 48(2):93–100.
- Agarwal I, Bauer AM, Jackman TR, Karanth KP. 2014. Insights into Himalayan biogeography from geckos: a molecular phylogeny of Cyrtodactylus (Squamata: Gekkonidae). Mol Phylogenet Evol. 80:145–155. doi:https://doi.org/10.1016/j.ympev.2014.07.018.
- Agarwal I, Khandekar A, Bauer AM. 2018c. A new bent-toed gecko (Squamata: Gekkonidae: Cyrtodactylus) from the Western Himalayas, Himachal Pradesh, India. Zootaxa. 4446(4):442–454. doi:https://doi.org/10.11646/zootaxa.4446.4.2.
- Agarwal I, Mahony S, Giri VB, Chaitanya R, Bauer AM. 2018a. Two new species of bent toed geckos, Cyrtodactylus Gray, 1827 (Squamata: Gekkonidae) from Northeast India with comments on name-bearing types from the region. Zootaxa. 4420(3):334–356. doi:https://doi.org/10.11646/zootaxa.4420.3.2.
- Agarwal I, Mahony S, Giri VB, Chaitanya R, Bauer AM. 2018b. Six new Cyrtodactylus (Squamata: Gekkonidae) from northeast India. Zootaxa. 4524(5):501–535. doi:https://doi.org/10.11646/zootaxa.4524.5.1.
- Ahsan MF. 1998. Herpetofauna of Bangladesh: present status, distribution and conservation. In: De Silva A, editor. Biology and conservation of the amphibians, reptiles and their habitats in South Asia. Gampola: Amphibia and Reptile Research Organization of Sri Lanka; p. 9–17.
- Al-Razi H, Maria M, Hasan S. 2018. First record of the recently described Cyrtodactylus tripuraensis Agarwal, Mahony, Giri, Chaitanya & Bauer, 2018 (Squamata, Gekkonidae) in Bangladesh. Check List. 14(6):1105–1108. doi:https://doi.org/10.15560/14.6.1105.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. doi:https://doi.org/10.1016/S0022-2836(05)80360-2.
- Annandale N. 1906. A new gecko from the Eastern Himalayas. J Asiatic Soc Bengal. 2(7):287–288.
- Annandale N. 1912. Zoological results of the Abor Expedition, 1911–1912. I. Batrachia. Rec Indian Mus 8(1):7–36 + pls. II–IV. doi:https://doi.org/10.5962/bhl.part.1186.
- Annandale N. 1913. The Indian geckos of the genus Gymnodactylus. Rec Indian Mus. 9:309–326.
- Barabanov AV, Doronin IV. 2020. Annotated list of amphibian and reptile taxa described by Ilya Sergeevich Darevsky (1924–2009). Zootaxa. 4803(1):152–168. doi:https://doi.org/10.11646/zootaxa.4803.1.8.
- Bauer AM. 2002. Two new species of Cyrtodactylus (Squamata: Gekkonidae) from Myanmar. Proc Calif Acad Sci. 53:73–86.
- Bauer AM. 2003. Descriptions of seven new Cyrtodactylus (Squamata: Gekkonidae) with a key to the species of Myanmar (Burma). Proc Calif Acad Sci. 54:463–498.
- Bauer AM, Masroor R, Titus-Mcquillan J, Heinicke MP, Daza JD, Jackman TR. 2013. A preliminary phylogeny of the Palearctic naked-toed geckos (Reptilia: Squamata: Gekkonidae) with taxonomic implications. Zootaxa. 3599(4):301–324. doi:https://doi.org/10.11646/zootaxa.3599.4.1.
- Bauer AM, Sumontha M, Pauwels OSG. 2003. Two new species of Cyrtodactylus (Reptilia: Squamata: Gekkonidae) from Thailand. Zootaxa. 376(1):1–18. doi:https://doi.org/10.11646/zootaxa.376.1.1.
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2017. GenBank. Nucleic Acids Res. 45(D1):D37–D42. doi:https://doi.org/10.1093/nar/gkw1070.
- Biju SD, Senevirathne G, Garg S, Mahony S, Kamei RG, Thomas A, Shouche Y, Raxworthy CJ, Meegaskumbura M, Van Bocxlaer I. 2016. Frankixalus, a new rhacophorid genus of tree hole breeding frogs with oophagous tadpoles. PLOS ONE. 11(1):e0145727. doi:https://doi.org/10.1371/journal.pone.0145727.
- Blyth E. 1861. Report of Curator, Zoological Department. J Proc Asiatic Soc Bengal. 29(1 “1860”):87–115.
- Boulenger GA. 1893. Concluding report on the reptiles and batrachians obtained in Burma by Signor L. Fea, dealing with the collection made in Pegu and the Karin Hills in 1887–8. Annali del Museo Civico di Storia Naturale di Genova, Series. 2(13):304–347 + Pls. VII–XII.
- Boulenger GA. 1905. XLVIII.—On some batrachians and reptiles from Tibet. Ann Mag Nat Hist Ser 7. 15(88):378–379. doi:https://doi.org/10.1080/03745480509443063.
- Brennan IG, Bauer AM, Van Tri N, Wang Y-y, Wang W-z, Zhang Y-p, Murphy RW. 2017. Barcoding utility in a mega-diverse, cross-continental genus: keeping pace with Cyrtodactylus geckos. Sci Rep. 7(1):1–11. doi:https://doi.org/10.1038/s41598-017-05261-9.
- Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2016. GenBank. Nucleic Acids Res. 44(D1):D67–D72. doi:https://doi.org/10.1093/nar/gkv1276.
- Darevsky IS, Helfenberger N, Orlov N, Shah K. 1998. Two new species of the genus Gonydactylus (Sauria: Gekkonidae) from eastern Nepal. Russ J Herpetol. 4(2 “1997”):89–93. doi:https://doi.org/10.30906/1026-2296-1997-4-2-89-93.
- Das A, Sharma P, Surendran H, Nath A, Ghosh S, Dutta D, Mondol J, Wangdi Y. 2016. Addition to the herpetofauna of Royal Manas National Park, Bhutan, with six new country records. Herpetol Notes. 9:261–278.
- Duda PL, Sahi DN. 1978. Cyrtodactylus himalayanus: a new gekkonid species from Jammu, India. J Herpetol. 12(3):351–354. doi:https://doi.org/10.2307/1563616.
- Edgar RC. 2004. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5(1):113. doi:https://doi.org/10.1186/1471-2105-5-113.
- Garg S, Das A, Kamei RG, Biju SD. 2018. Delineating Microhyla ornata (Anura, Microhylidae): mitochondrial DNA barcodes resolve century-old taxonomic misidentification. Mitochondrial DNA Part B. 3(2):856–861. doi:https://doi.org/10.1080/23802359.2018.1501286.
- Giri VB, Chaitanya R, Mahony S, Lalrounga S, Lalrinchhana C, Das A, Sarkar V, Karanth P, Deepak V. 2019a. On the systematic status of the genus Oriocalotes Günther, 1864 (Squamata: Agamidae: Draconinae) with the description of a new species from Mizoram state, Northeast India. Zootaxa. 4638(4):451–484. doi:https://doi.org/10.11646/zootaxa.4638.4.1.
- Giri VB, Gower DJ, Das A, Lalremsanga HT, Lalronunga S, Captain A, Deepak V. 2019b. A new genus and species of natricine snake from northeast India. Zootaxa. 4603(2):241–264. doi:https://doi.org/10.11646/zootaxa.4603.2.2.
- Gray JE. 1827. A synopsis of the genera of saurian reptiles, in which some new genera are indicated, and the others reviewed by actual examination. Philos Mag Ann Chem Math Astron Nat Hist Gen Sci. 2:54–58. doi:https://doi.org/10.1080/14786442708675620.
- Grismer LL, Wood Jr. PL, Poyarkov NA, Le MD, Kraus F, Agarwal I, Oliver PM, Nguyen SN, Nguyen TQ, Karunarathna S, et al. 2021. Phylogenetic partitioning of the third-largest vertebrate genus in the world, Cyrtodactylus Gray, 1827 (Reptilia; Squamata; Gekkonidae) and its relevance to taxonomy and conservation. Vertebrate Zool. 71:101–154.
- Grismer LL, Wood Jr. PL, Quah ESH, Murdoch ML, Grismer MS, Herr MW, Espinoza RE, Brown RM, Lin A. 2018d. A phylogenetic taxonomy of the Cyrtodactylus peguensis group (Reptilia: Squamata: Gekkonidae) with descriptions of two new species from Myanmar. PeerJ. 6:e5575. doi:https://doi.org/10.7717/peerj.5575.
- Grismer LL, Wood Jr. PL, Quah ESH, Thura MK. 2020. Origin, diversity, and conservation of karst-associated Bent-toed geckos (genus Cyrtodactylus) in Myanmar (Burma). Isr J Ecol Evol. 1(aop):1–7. doi:https://doi.org/10.1163/22244662-20191094.
- Grismer LL, Wood Jr. PL, Quah ESH, Thura MK, Herr MW, Lin AK. 2019b. A new species of forest-dwelling Cyrtodactylus Gray (Squamata: Gekkonidae) from the Indawgyi Wildlife Sanctuary, Kachin State, Myanmar. Zootaxa. 4623(1):1–25. doi:https://doi.org/10.11646/zootaxa.4623.1.1.
- Grismer LL, Wood Jr. PL, Thura MK, Quah ESH, Murdoch ML, Grismer MS, Herr MW, Lin A, Kyaw H. 2018b. Three more new species of Cyrtodactylus (Squamata: Gekkonidae) from the Salween Basin of eastern Myanmar underscore the urgent need for the conservation of karst habitats. J Nat Hist. 52(19–20):1243–1294. doi:https://doi.org/10.1080/00222933.2018.1449911.
- Grismer LL, Wood Jr. PL, Thura MK, Win NM, Grismer MS, Trueblood LA, Quah ESH. 2018c. A re-description of Cyrtodactylus chrysopylos Bauer (Squamata: Gekkonidae) with comments on the adaptive significance of orange coloration in hatchlings and descriptions of two new species from eastern Myanmar (Burma). Zootaxa. 4527(2):151–185. doi:https://doi.org/10.11646/zootaxa.4527.2.1.
- Grismer LL, Wood Jr. PL, Thura MK, Win NM, Quah ESH. 2019a. Two more new species of the Cyrtodactylus peguensis group (Squamata: Gekkonidae) from the fringes of the Ayeyarwady Basin, Myanmar. Zootaxa. 4577(2):274–294. doi:https://doi.org/10.11646/zootaxa.4577.2.3.
- Grismer LL, Wood Jr. PL, Thura MK, Zin T, Quah ESH, Murdoch ML, Grismer MS, Lin A, Kyaw H, Lwin N. 2018a. Twelve new species of Cyrtodactylus Gray (Squamata: Gekkonidae) from isolated limestone habitats in east-central and southern Myanmar demonstrate high localized diversity and unprecedented microendemism. Zool J Linn Soc. 182(4):862–959. doi:https://doi.org/10.1093/zoolinnean/zlx057.
- Harvey MB, O’Connell KA, Barraza G, Riyanto A, Kurniawan N, Smith EN. 2015. Two new species of Cyrtodactylus (Squamata: Gekkonidae) from the southern Bukit Barisan Range of Sumatra and an estimation of their phylogeny. Zootaxa. 4020(3):495–516. doi:https://doi.org/10.11646/zootaxa.4020.3.5.
- Harvey MB, O’Connell KA, Wostl E, Riyanto A, Kurniawan N, Smith EN, Grismer LL. 2016. Redescription Cyrtodactylus lateralis (Werner) (Squamata: Gekkonidae) and phylogeny of the prehensile-tailed Cyrtodactylus. Zootaxa. 4107(4):517–540. doi:https://doi.org/10.11646/zootaxa.4107.4.3.
- Jerdon TC. (1870) Notes on Indian herpetology. Proc Asiatic Soc Bengal. 1870:66–85.
- Johnson CB, Quah ESH, Anuar S, Muin MA, Wood Jr. PL, Grismer JL, Greer LF, Chan KO, Ahmad N, Bauer AM, et al. 2012. Phylogeography, geographic variation, and taxonomy of the Bent-toed Gecko Cyrtodactylus quadrivirgatus Taylor, 1962 from Peninsular Malaysia with the description of a new swamp dwelling species. Zootaxa. 3406:39–58. doi:https://doi.org/10.11646/zootaxa.3406.1.3.
- Kamei RG. 2017. An overview of caecilians (Amphibia: Gymnophiona) of North East India. In: Das A, editor. Diversity and Ecology of Amphibians of India. ENVIS Bulletin: Wildlife and Protected Areas (Vol. 19). Dehradun: Wildlife Institute of India; p. 49–72.
- Kamei RG, Gower DJ, Wilkinson M, Biju SD. 2013. Systematics of the caecilian family Chikilidae (Amphibia: Gymnophiona) with the description of three new species of Chikila from northeast India. Zootaxa. 3666(4):401–435. doi:https://doi.org/10.11646/zootaxa.3666.4.1.
- Kamei RG, Mahony S. 2021. A new species of Bent-toed gecko (Squamata: Gekkonidae: Cyrtodactylus Gray, 1827) from the Garo Hills, Meghalaya State, north-east India, and discussion of morphological variation for C. urbanus. Herpetol J. 31(3):177–196. doi:https://doi.org/10.33256/31.3.177196.
- Kamei RG, Wilkinson M, Gower DJ, Biju SD. 2009. Three new species of striped Ichthyophis (Amphibia: Gymnophiona: Ichthyophiidae) from the northeast Indian states of Manipur and Nagaland. Zootaxa. 2267(1):26–42. doi:https://doi.org/10.11646/zootaxa.2267.1.2.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:https://doi.org/10.1093/bioinformatics/bts199.
- Khan MS. 1993. A new angular-toed gecko from Pakistan, with remarks on the taxonomy and a key to the species belonging to genus Cyrtodactylus (Reptilia: Sauria: Gekkonidae). Pak J Zool. 25(1):67–73.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:https://doi.org/10.1093/molbev/msw054.
- Li P-P. 2007. Description of a new subspecies of Cyrtodactylus khasiensis from China. Acta Zootaxonomica Sinica. 32:733–737.
- Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ. 1997. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol. 14(1):91–104. doi:https://doi.org/10.1093/oxfordjournals.molbev.a025706.
- Mahony S. 2009. Taxonomic status of Cyrtodactylus khasiensis tamaiensis (Smith, 1940) and description of a new species allied to C. chrysopylos Bauer, 2003 from Myanmar (Reptilia: Gekkonidae). Hamadryad. 34(1):62–74.
- Mahony S, Ahmed M, Hossain MK, Kabir MM, Hasan MK. 2009b. Cyrtodactylus ayeyarwadyensis Bauer, 2003 (Reptilia: Gekkonidae) in Bangladesh with habitat details of new collection localities and a discussion of morphological variation. Salamandra. 45(4):245–250.
- Mahony S, Reza AHMA. 2008. A herpetofaunal collection from the Chittagong Hill tracts, Bangladesh, with two new species records for the country. Hamadryad. 32(1):45–56.
- Mahony S, Foley NM, Biju SD, Teeling EC. 2017. Evolutionary history of the Asian horned frogs (Megophryinae): integrative approaches to timetree dating in the absence of a fossil record. Mol Biol Evol. 34(4):744–771 + S.I. 38 p. doi:https://doi.org/10.1093/molbev/msw267.
- Mahony S, Hasan MK, Kabir MM, Ahmed M, Hossain MK. 2009a. A catalogue of herpetofauna in the collection of Jahangirnagar University, Dhaka, Bangladesh. Hamadryad. 34(1):80–94.
- Mahony S, Kamei RG, Teeling EC, Biju SD. 2018. Cryptic diversity within the Megophrys major species group (Amphibia: Megophryidae) of the Asian horned frogs: phylogenetic perspectives and a taxonomic revision of South Asian taxa, with descriptions of four new species. Zootaxa. 4523(1):1–96. doi:https://doi.org/10.11646/zootaxa.4523.1.1.
- Mahony S, Kamei RG, Teeling EC, Biju SD. 2020. Taxonomic review of the Asian Horned Frogs (Amphibia: Megophrys Kuhl & Van Hasselt) of Northeast India and Bangladesh previously misidentified as M. parva (Boulenger), with descriptions of three new species. J Nat Hist. 54(1–4):119–194. doi:https://doi.org/10.1080/00222933.2020.1736679.
- Mathew R. 2005. Reptilia: Squamata. In: Director, editor. Fauna of Manipur, Part I (Vertebrates and Animal Fossils). State Fauna Series 10. Kolkata: Zoological Survey of India; p. 119–122.
- Mathew R, Sen N. 2009. Studies on little known amphibian species of North East India. Zool Surv India Occasional Pap. 293:1–64.
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Institute of Electrical and Electronics Engineers, editor. Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans: Piscataway; p. 1–8. doi:https://doi.org/10.1109/GCE.2010.5676129.
- Mirza ZA, Bhosale H, Ansari F, Phansalkar P, Sawant M, Gowande G, Patel H. 2021. A new species of geckos of the genus Cyrtodactylus Gray, 1827 from Arunachal Pradesh, India. Evol Syst. 5(1):13–23. doi:https://doi.org/10.3897/evolsyst.5.61667.
- Mittermeier RA, Gill PR, Hoffman M, Pilgrim J, Brooks T, Mittermeier CG, Lamoreux J, Da Fonseca GAB. (2004) Hotspots revisited: earth’s biologically richest and most endangered terrestrial ecoregions. Cemex, Conservation International and Agrupacin Sierra Madre, Monterrey, Mexico, 431 pp.
- Muansanga L, Decemson H, Biakzuala L, Hmar GZ, Lalremsanga HT, Das M, Purkayastha J. 2020. First record of the Jampui bent-toed gecko, Cyrtodactylus montanus Agarwal, Mahony, Giri, Chaitanya, and Bauer 2018 (Squamata: Gekkonidae), from Mizoram, India. ICRF Reptiles Amphibians Conserv Nat Hist. 27(2):267–268. doi:https://doi.org/10.17161/randa.v27i2.14325.
- Murthy TSN, Singh KS. 1982. Notes on some lizards from Imphal, Manipur. Geobios New Rep. 1(2):140–142.
- Ningobam B, Bordoloi S. 2007. Amphibian fauna of Loktak Lake, Manipur, India, with ten new records for the state. Zoos’ Print J. 22(5):2688–2690. doi:https://doi.org/10.11609/JoTT.ZPJ.1557.2688-90.
- Pawar S, Birand A. (2001) A survey of amphibians, reptiles, and birds of Northeast India. CERC Technical Report #6, Center for Ecological Research and Conservation, Mysore, 75 pp.
- Purkayastha J, Das M, Bohra SC, Bauer AM, Agarwal I. 2020. Another new Cyrtodactylus (Squamata: Gekkonidae) from Guwahati, Assam, India. Zootaxa. 4732(3):375–392. doi:https://doi.org/10.11646/zootaxa.4732.3.2.
- Purkayastha J, Lalremsanga HT, Bohra SC, Biakzuala L, Decemson HT, Muansanga L, Vabeiryureilai M, Chauhan S, Rathee YS. 2021. Four new Bent-toed geckos (Cyrtodactylus Gray: Squamata: Gekkonidae) from northeast India. Zootaxa. 4980(3):451–489. doi:https://doi.org/10.11646/zootaxa.4980.3.2.
- Rambaut A. 2009. FigTree version 1.3.1. Computer program distributed by the author. [accessed 2012 Jan]. http://tree.bio.ed.ac.uk/software/figtree/
- Riyanto A, Farajallah A, Hamidy A, Fitriana YS, Munir M, Kurniawan N, Smith EN. 2020. Taxonomic evaluation of two similar bent-toed geckos Squamata: Gekkonidae: Cyrtodactylus Gray, 1827) from East Java, Indonesia. Zootaxa. 4830(1):186–196. doi:https://doi.org/10.11646/zootaxa.4830.1.8.
- Sarkar AK, Chandra PK, Ray S. 2005. Amphibia. In: Director, editor. Fauna of Manipur, Part I (Vertebrates and Animal Fossils). State Fauna Series 10. Kolkata: Zoological Survey of India; p. 123–132.
- Schlegel H. 1837. Abbildungen neuer oder unvollständig bekannter Amphibien, nach der Natur oder dem Leben entworfen. Düsseldorf: Arnz & Comp; p. i–xiv + 141. [+ pls. 1–50: 1837–1844].
- Siler CD, Oaks JR, Esselstyn JA, Diesmos AC, Brown RM. 2010. Phylogeny and biogeography of Philippine bent-toed geckos (Gekkonidae: Cyrtodactylus) contradict a prevailing model of Pleistocene diversification. Mol Phylogenet Evol. 55(2):699–710. doi:https://doi.org/10.1016/j.ympev.2010.01.027.
- Singh KHS. 1977. Newts of Manipur. Sci Rep. 14(2):721–722.
- Smith MA. 1935. The Fauna of British India, including Ceylon and Burma. Reptiles and Amphibia. Vol. II. Sauria. London: Taylor and Francis; p. xiii + 440 + pl. 1 + 2 maps.
- Smith MA. 1940. The amphibians and reptiles obtained by Mr. Ronald Kaulback in Upper Burma. Rec Indian Mus. 42:465–486.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:https://doi.org/10.1093/bioinformatics/btu033.
- Steindachner F. 1867. Reptilien. In: Anonymous, editor. Reise der Österreichischen Fregatte Novara um die Erde in den Jahren 1857, 1858, 1859 unter den Befehlen des Commodore B. von Wüllerstorf-Urbair (Zoologie), Vol. 1, Part 3. 1869 on title page. Wien: K. Gerold’s Sohn/Kaiserlich-Königl. Hof- und Staatsdruckerei; p. 1–98 + 3 pls.
- Stoliczka F. 1871. Notes on new or little known Indian lizards. Proc Asiatic Soc Bengal. 1871(September):192–195.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526. doi:https://doi.org/10.1093/oxfordjournals.molbev.a040023.
- Uetz P. 2021. The Reptile Database. [accessed 2020 Dec 11]. http://reptile-database.reptarium.cz
- Wood Jr PL, Heinicke MP, Jackman TR, Bauer AM. 2012. Phylogeny of bent-toed geckos (Cyrtodactylus) reveals a west to east pattern of diversification. Mol Phylogenet Evol. 65(3):992–1003. doi:https://doi.org/10.1016/j.ympev.2012.08.025.
Appendix I:
Comparative specimens examined
Cyrtodactylus himalayicus: holotype: male (ZSIK 15716), from ‘Kurseong, Darjeeling Dist. (5,000 ft)’ [= Kurseong Subdivision (ca. 1524 m asl.), Darjeeling District, West Bengal State, India]; referred material: adult female (ZSIK 19546), from ‘Gopaldhara, Darjeeling Dist’. [=Gopaldhara Tea Estate, near Mirik Town, Darjeeling District, West Bengal State, India]. Cyrtodactylus mandalayensis: holotype: subadult male (BMNH 1900.9.20.1), from Mogok Town (ca. 22.922896, 96.506150, ca. 1170 m asl.) or its vicinity, Mogok Township, Pyin Oo Lwin District, Mandalay Region, Myanmar. Cyrtodactylus tamaiensis: holotype: adult male (BMNH 1946.823.22), from ‘Pangnamdim, Nam Tamai Valley, Burma’ [= Pangnamdim, Kachin State, Myanmar].
Appendix II.
Table of localities and GPS coordinates plotted in . Taxa marked with asterisks indicate referred localities; remaining localities are type localities.
