ABSTRACT
A new species of Delorhachis Karsch, 1896, Delorhachis nouabaleensis sp. n. is described from the Nouabalé-Ndoki National Park in northern Republic of Congo based on morphological and genetic evidence. This area is likely to have high levels of endemism due to the surrounding Sangha River Interval.
http://www.zoobank.org/urn:lsid:zoobank.org:pub:CBAF262B-4616-4E45-9293-35DE836972D9
KEYWORDS:
Introduction
The Afrotropical limacodid genus Delorhachis Karsch, 1896 currently contains 20 species and one subspecies distributed throughout much of sub-Saharan Africa. Members of the genus are distinctive and can easily be distinguished by the presence of a medio-ventral fore wing fascia and dark venation in the distal half of the fore wing. The genus was comprehensively revised by Taberer et al. (Citation2023), who described several new species and assigned each taxon to one of five species groups. A recent expedition to the Nouabalé-Ndoki National Park in northern Republic of Congo by the African Natural History Research Trust (ANHRT) revealed the presence of a peculiar Delorhachis species not matching any known species but sharing affinities with members of several species groups. Upon closer examination of external and genital morphology, as well as DNA sequencing of the COI barcode region, its specific distinctness was discerned and it is described herein as a new species: Delorhachis nouabaleensis sp. n.
Material and methods
Images of adults were taken using a Canon EOS 650d DSLR camera equipped with a Sigma DG Macro 105 mm 1:2.8 lens. The genitalia were dissected and stained with Eosin Y, applying standard methods of preparation (Lafontaine and Mikkola Citation1987), then embedded in Euparal on microscope slides. The genitalia preparations were photographed using a Canon EOS 700D camera mounted on a Leitz Diaplan compound microscope. Label information provided in quotation marks is transcribed verbatim. A new line is denoted with ‘|’ and a different label with ‘||’.
DNA extraction and sequencing was undertaken at the Natural History Museum, London, UK. DNA was extracted using a Qiagen DNeasy Blood and Tissue Kit with ‘non-destructive’ modifications as described in Polaszek et al. (Citation2013). An entire hind leg was lysed for 8 h at 55°C in a water bath, without grinding or other damage. After removal of the lysate, the extracted leg was washed in distilled water, and prepared for eventual reuniting with the adult specimen from which it had been removed. The lysate was cleaned using the standard DNeasy spin-column protocol, with final elution of DNA into 100 uL of Qiagen Elution Buffer. A 658 bp region of the COI gene was PCR amplified using universal Folmer primers, and Polymerase chain Reactions (PCR) were conducted using the KAPA 3 G Plant PCR kit with 4.8 µL DNA extract. Subsequently, COI amplicons were combined with other uniquely tagged samples, and a sequencing library was constructed using the V14 Ligation Sequencing Kit (SQK-LSK114) from Oxford Nanopore Technologies. Sequencing was performed on a GridION instrument using a v. 10.4.1 Flongle. For data analysis, ONTBarcoder v. 0.1.9 was employed for demultiplexing, alignment, consensus creation, and barcode polishing. The resulting barcodes were compared against the BOLD database using BOLDigger v. 2.1.1 for taxonomic identification.
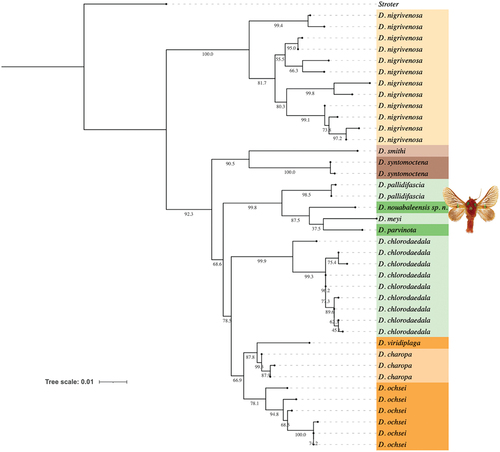
The COI sequence of the holotype of D. nouabaleensis was compared with other Delorhachis sequences as detailed in Taberer et al. (Citation2023) (n = 39). The outgroup was a member of the genus Stroter Karsch, 1899, another Afrotropical limacodid. Sequences were aligned using MUSCLE in MEGA v. X (Kumar et al. Citation2018) and pairwise distances within and between species were calculated using the Kimura two-parameter model (Kimura Citation1980). Phylogenetic tree searches were performed using Bayesian inference (BI) and maximum likelihood (ML). BI analyses were performed using MrBayes v. 3.2.7a (Ronquist et al. Citation2012). Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses were run with four chains (one cold and three heated) for 10,000,000 generations sampling every 100 generations, discarding the first 25% as burn-in. The two runs converged with the standard deviation of split frequencies <0.001. ML analyses were performed on raxmlGUI 2.0 (Edler et al. Citation2021) using RAxML-NG (Kozlov et al. Citation2019) with 1000 repetitions and a GTR+FO+G4m model. Support for clades was evaluated for BI using posterior probabilities and ML using non-parametric bootstrapping. The ML phylogeny was visualised and annotated in iTol v. 6.9 (Letunic and Bork Citation2006) and Procreate, and is shown in .
Delorhachis nouabaleensis sp. n.
Holotype
Male, “REPUBLIC OF CONGO 341m | Sangha Prov., Nouabale-Ndoki | National Park, Bomassa camp | (Secondary forest) | 02º12’36.9”N, 16º11’30.2”E | 16–23.ix.2022 MV Light Trap | Dérozier, V., Fouka, B., | Kirk-Spriggs, A., Takano, H. Leg. | ANHRT:2022.14” || “ANHRTUK | 00293369” || “Gen. slide No. | TT 223 | prep. by T. R. Taberer”
Description
Male
External morphology
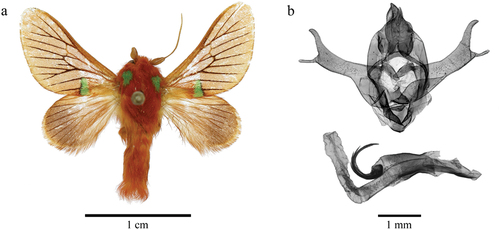
()) Fore wing length 11 mm. Head, collar, tegula and thorax vermillion, antenna brown, bipectinate in basal fourth, otherwise filiform. Tegula with one green rounded patch. Dorsal side of thorax vermillion; ventral side pale beige without markings. Legs vermillion laterally, beige medially; tibial spur formula 0-0-0. Abdomen vermillion dorsally, pale beige ventrally. Fore wing upperside: Short, broad, triangular, with rounded apex; ground colour vermillion, veins contrasting black in postmedial area. Medio-ventral fascia Veronese green, narrow, rectangular, edged with black distally. Fringe short, slightly darker than ground colour. Hind wing upperside: Proximal half beige, distal half pale brown with weakly defined dark brown veins postmedially. Fringe long, pale brown. Fore wing underside: Pale vermillion without markings. Hind wing underside: Pale beige in proximal half and pale brown in distal half; fringe same as ground colour.
Genitalia
()) Uncus short, tapered, apically pointed. Tegumen broad. Gnathos broad, rounded with a pointed apical process. Juxta rounded basally, with two very short, ventral, ribbon-like distal processes; manica with a bunch of moderately short sclerotised, curved pseudocornuti. Vinculum short and broad, rounded. Valva broad at base, slightly constricted medially, distally forming a long, narrow, slightly curved, dorso-apical process with a rounded point and a similarly long but narrower, distally rounded ventral process. Phallus relatively short, straight with sclerotised, apically pointed ventral surface in its basal half. Vesica membranous without cornuti.
Female
Unknown.
Diagnosis
Despite its phenotypic similarities to members of several species groups, Delorhachis nouabaleensis is here considered to belong to the D. chlorodaedala species group. In morphology, the closest relative of D. nouabaleensis is Delorhachis tommasoi Giusti, Taberer, Fiebig and László, 2023, with both species exhibiting the small green medio-ventral fascia on the fore wing, which in the new species is rectangular whilst in D. tommasoi it is rounded. In the ML phylogeny (), D. nouabaleensis was recovered within the cluster containing Delorhachis pallidifascia Taberer, Fiebig, Giusti and László, Citation2023, Delorhachis meyi Taberer, Fiebig, Giusti and László, Citation2023 and Delorhachis parvinota Taberer, Fiebig, Giusti and László, Citation2023 with high bootstrap support (99.8%); all of these species are members of the D. chlorodaedala species group (see Taberer et al. Citation2023 for further discussions on the phylogenetic analyses within this genus). The new species was recovered as sister to the cluster containing D. meyi (pairwise distance: 2.02%) and D. parvinota (pairwise distance: 2.45%) (note: D. tommasoi currently does not have COI sequence data). It is interesting to note, however, that the single holotype specimen lacks the central green patch on the thorax typical of all other members of the D. chlorodaedala species group, which suggests this may not be a synapomorphic character (as listed in table 1 of Taberer et al. Citation2023).
Figure 2. Phylogenetic tree (maximum likelihood model) of the genus Delorhachis, highlighting recovery of the new species Delorhachis nouabaleensis sp. n.
The hind wings of D. nouabaleensis are dissimilar to others in the D. chlorodaedala species group, and are pale brown with weakly defined brown veins in the distal portion as opposed to the typical uniform pale beige. This is similar to some members of the D. syntomoctena species group, such as D. syntomoctena (Tams, 1929) and D. nimbaensis Taberer, Fiebig, Giusti and László, Citation2023, clearly demonstrating its unique characteristics. Interestingly, the tibial spur formula of this species is 0-0-0, similar to the D. syntomoctena species group and not the D. chlorodaedala species group, whose members have a formula of 0-2-2. Finally, the dorsal side of the abdomen in the new species is covered in vermillion hair scales, which are generally only present in the first two segments in both the D. viridiplaga and D. chlorodaedala species groups.
When considering features of the male genitalia, however, these strongly suggest that D. nouabaleensis belongs to the D. chlorodaedala species group (for example, the bilobate valva, the relatively straight phallus and the cluster of curled pseudocornuti; see table 1 of Taberer et al. Citation2023). The gnathos of the new species bears a close similarity to that of D. pallidifascia Taberer, Fiebig, Giusti and László, Citation2023, a West African species with a pale green fore wing fascia, whilst the distal processes of the valva are most reminiscent of D. tommasoi and D. mariae Dufrane, 1945 (two other members of the D. chlorodaedala species group). However, both distal processes of the valva in the new species are notably longer and thinner than those of either of its two phylogenetically closest congeners, D. meyi and D. parvinota, most noticeably in the ventral process.
Etymology
This new species is named after its type locality of Nouabalé-Ndoki National Park, Republic of Congo.
Distribution and biogeography
Delorhachis nouabaleensis is so far known from only a single specimen from the type locality within the Congo Basin lowland forests of northern Republic of Congo. Nouabalé-Ndoki National Park is situated within the Sangha River Interval, a region that has experienced major vegetation changes and been greatly impacted by climate changes in the past (Gond et al. Citation2013). An increasing amount of data seems to suggest that this area harbours endemic or near-endemic lepidopteran species with close affinities to coastal Cameroon and Gabon, as well as to the forests east of the Congo River (H. Takano, in prep.), and in the case of D. nouabaleensis, it may have evolved in allopatry across the Congo River from the closely related D. tommasoi, the latter’s most westerly record being Mbandaka, Democratic Republic of the Congo (as Coquilhatville in Taberer et al. Citation2023, p. 77).
Acknowledgements
The Wildlife Conservation Society, Congo and their staff are thanked for authorising research in Nouabalé-Ndoki National Park, in particular Richard Malonga (Director, Republic of Congo Program), Morgane Cournarie (Program Coordinator, Republic of Congo Program), Ben Evans (Park Director, Nouabalé-Ndoki), Vittoria Estienne (Director of Research and Biomonitoring), Onesi Samba (Research Assistant) and Yako Valentin (Administrative Manager). Joseph Goma-Tchimbakala, (Director General, Institut National de Recherche en Sciences Exactes et Naturelles (IRSEN)) and Jean Bosco Nganongo (Director, Direction de la Faune et des aires protégées (DFAP)) are gratefully acknowledged for the seamless collaboration in issuing the necessary research and export permits; and Victor Mamonekene (IRSEN and Marien Ngouabi University) is thanked for his valuable assistance during fieldwork. I am grateful to Hitoshi Takano (ANHRT) for useful discussions on the biogeography of this region. Finally, I extend my sincere thanks to Andrew Polaszek and Jordan Beasley (NHMUK) for extracting and sequencing the COI barcode of the holotype of this species, allowing for its inclusion in the Delorhachis phylogeny.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Edler D, Klein J, Antonelli A, Silvestro D. 2021. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol. 12:373–377. doi: 10.1111/2041-210X.13512.
- Gond V, Fayolle A, Pennec A, Cornu G, Mayaux P, Camberlin P, Doumenge C, Fauvet N, Gourlet-Fleury S. 2013. Vegetation structure and greenness in Central Africa from Modis multi-temporal data. Philos Trans R Soc B. 368:20120309. doi: 10.1098/rstb.2012.0309.
- Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120. doi: 10.1007/BF01731581.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455. doi: 10.1093/bioinformatics/btz305.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Lafontaine JD, Mikkola K. 1987. Lock–and–key system in the inner genitalia of Noctuidae (Lepidoptera) as a taxonomic character. Entomologiske Meddelelser. 55:161–167.
- Letunic I, Bork P. 2006. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 23(1):127–128. doi: 10.1093/bioinformatics/btl529.
- Polaszek A, Ayshford T, Yahya BE, Fusu L. 2013. Wallaceaphytis: an unusual new genus of parasitoid wasp (Hymenoptera: Aphelinidae) from Borneo. J Nat History. 48:1111–1123. doi: 10.1080/00222933.2013.852264.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Taberer TR, Fiebig R, Giusti A, László G. 2023. Taxonomic revision of the genus Delorhachis Karsch 1896 (Lepidoptera: Limacodidae). J Nat History. 57(1–4):54–129. doi: 10.1080/00222933.2022.2157346.