?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
We present the computer program DeCE developed at Los Alamos National Laboratory, which is open-source software to assist in producing evaluated nuclear data files. DeCE manipulates a provided evaluated nuclear data file interactively, then creates a new file in an ENDF-6 legitimate format. DeCE also extracts numerical data given in an evaluated file and converts into a more readable format. The code includes two C++ libraries, ENDFLIB and ENDFIO, which fulfill a demand for easy interface between a modern computer programming technique and the inflexible ENDF-6 format. We overview the DeCE code and the libraries employed there, and demonstrate their capabilities.
1. Introduction
The evaluated nuclear data libraries, such as JENDL-4.0 [Citation1], ENDF/B-VIII.0 [Citation2], JEFF-3 [Citation3], and CENDL-3 [Citation4] are the indispensable resource for nuclear technology whenever the particle transport simulation is involved. These libraries are revised occasionally by including advanced experimental and theoretical knowledge of nuclear reaction and structure, as well as an innovative method to store important information of nuclear reaction mechanisms in an ASCII text file. For example, when one looks into the older version of the evaluated libraries, secondary particle energy and angular distributions are often stored separately in a data file, namely the angular distribution is in (
stands for the file number in the ENDF-6 format [Citation5]) and the energy distribution can be found in
. Such representation of the secondary particle energy and angular distributions is regarded as obsolete, especially insufficient for fusion technology applications, and modern evaluated libraries store such information in
: the energy and angle correlated distribution. Note that we refer ENDF-6 as a data format that is designed to maintain the evaluated nuclear data in a computer file, and the readers shouldn’t be confused by the ENDF library.
Keeping the evaluated nuclear data library updated satisfies users’ demand for reducing uncertainties in radiation transport simulations owing to the evaluated nuclear data themselves. Having said that, production of the brand-new evaluated data file always creates another level of difficulties. This is because the ENDF-6 format [Citation5] is a rigorous protocol to transfer the information at data evaluation to nuclear applications, and one has to obey these inflexible rules even if ENDF-6 still persists an old-fashioned 80-column card-image.
Naturally, there exists a new proposal to use more flexible data formats, such as XML (eXtensible Markup Language), to maintain the nuclear reaction and structure data [Citation6]. However, unfortunately, a majority of computer programs still stick to the ENDF-6 format for the backward compatibility and quality assurance. In fact, the Generalized Nuclear Data Structure, GNDS Citation[6], retains ENDF-6 file reproducibility.
Nuclear data compilation into the standard ENDF-6 formatted file remains the mainstream in the nuclear data application field. Nakagawa developed the CRECTJ5 and CRECTJ6 codes [Citation7] for compiling the JENDL libraries [Citation1], taking advantage of implicit affinity between ENDF-6 and FORTRAN. Since recent codes for calculating nuclear reactions or processing nuclear data libraries, such as CCONE [Citation8], AMUR [Citation9], CoH3 [Citation10], and FRENDY [Citation11], are often written in C++, we anticipate that demand for a substantial interface between a modern computer programming technique and the stiff FORTRAN-ish rules does not vanish.
The computer program DeCE, ‘Descriptive Correction of ENDF-6 format,’ is open-source software developed at Los Alamos National Laboratory, to provide the interface between ENDF-6 and C++. DeCE facilitates manipulating the evaluated nuclear data files, as well as making a new evaluation in the legitimate ENDF-6 format. The code is built on top of core C++ libraries, ENDFLIB and ENDFIO, which offer an easy access to the past ENDF-6 formatted files. This paper presents the computational technique employed in DeCE and demonstrates how DeCE can promote the data science in the nuclear technology field. DeCE is a modest size C++ code, 1.2 MBytes, 20,000 lines, and available at GitHub (https://github.com/toshihikokawano/DeCE). DeCE runs on a UNIX-like platform, such as Linux and MacOS. At this moment Windows is not supported, but we plan to provide the Windows version as we anticipate there could be such a demand.
2. Object-oriented programming for ENDF-6 data
2.1. ENDF class
An evaluated nuclear data file consists of several parts, called the file number , where for example,
is for a general description,
is for resonance parameters,
is for reaction cross sections, and so on. Each
is also divided into smaller sections, identified by the reaction type number
;
is the total cross section, 2 is the elastic scattering, 102 is the capture, etc. We process the evaluated nuclear data section-by-section, which is specified by a pair of
numbers.
Although ENDF-6 is still in the old card-image, its well-structured design is suitable for converting data into a so-called object. The most primitive data structure defined in ENDF-6 is the CONT (control) record, which contains a couple of floating-point variables and four integers. This CONT line is copied into the CONT class of C++,
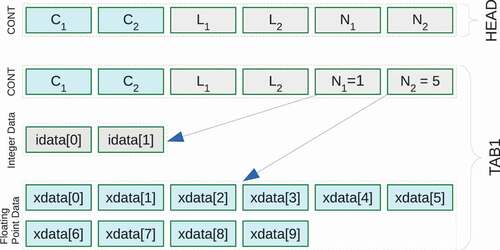
where we follow the ENDF-6 standard nomenclature. Each data section starts with a special CONT record, called HEAD. The HEAD record is followed by other data blocks whose data structure depends on the properties of stored nuclear data. For example, the cross-section data in are a set of incident particle energies and cross sections
with some integers to indicate how these data points are interpolated. ENDF-6 holds these data in one CONT as well as arrays for both integer and floating-point numbers, which is called a TAB1 record. We handle these HEAD + TAB1 data as a data block, which is shown in . In the case of scattering angular distribution, it consists of three-dimensional data (
,
,
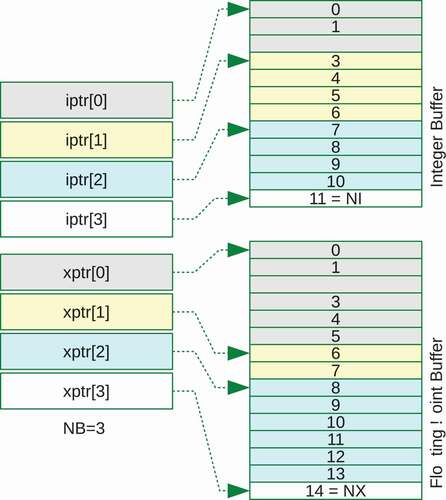
) that require more data blocks. Generally, since we don’t know how many CONT records we need, nor how large these data arrays could be, we define the ENDF class by using pointers,
Figure 1. An example of data block that contains the HEAD and TAB1 records defined in ENDF-6. This data block includes two CONT records, arrays of integer and floating-point numbers. indicates there are a pair of integer data, and
means there are five
pairs.
where is the HEAD record, the data are stored in integer and floating-point buffers, I and X, dynamically allocated in a heap.
and
are the pointer-to-pointers, where
contains the address of
-th element of
. The private member variable
is the number of data blocks currently stored in the ENDF object. shows how the I and X buffers are accessed by
and
. There are three data blocks, and the first block contains 3 integers and 6 floating-point numbers. The second block has 4 and 2, and so on. We do not memorize the total numbers of integer and floating-point data,
and
, because they can be calculated as
Figure 2. An integer pointer and a floating-point pointer to store arbitrary number of data in one-dimensional buffers. The last pointers point just one element outside the stored data.
so that we just need only. Similarly, the number of elements in each block can be calculated by the pointers.
2.2. ENDFLIB and ENDFIO libraries
In ENDF-6, data given in an section are organized by four data structures [Citation5], CONT, LIST, TAB1, and TAB2. The TAB2 block can contain other data blocks of LIST or TAB1. The ENDFLIB library includes many utilities to facilitate reading and writing these defined structures. For example, ENDFReadTAB1 is a function to read the TAB1-type data, ENDFWriteCONT prints a CONT record in the object currently accessing to.
Since ENDF-6 economizes on ASCII text field, a numeric text like ‘1.000000 + 3’, which is red easily by FORTRAN as , is preferred, while International Organization for Standardization, ISO 6093 – Representation of numerical values in character strings for information interchange – may require a letter E before the plus sign. The E-letter padding is automatically done in ENDFLIB.
The ENDFIO library offers an easy access to the evaluated nuclear data file. The function ENDFRead copies data in the file specified by to the object. The ENDF object data are printed by a function ENDFWrite in the ENDF-6 appropriate format. Since each section has a different data structure depending on
, one has to consult the ENDF-6 manual [Citation5] for extracting requisite information if needed. Otherwise, these functions, ENDFRead and ENDFWrite, should work properly for all the
sections.
2.3. ENDFDict class
The ENDFDict class controls the data structure in a given ENDF-6 file, as well as methods to access some information given in the header part of the evaluated data file. For instance, the member function getEMAX() tells the highest energy of the current file. Unlike the ENDF class, this object does not contain any nuclear data but plays a central role in making a new evaluated data file.
2.4. The DeCE code
By applying the programming technique employed in ENDF and ENDFDict classes, the computer code DeCE processes the evaluated nuclear data files interactively. DeCE reads and writes the ENDF-6 files through the ENDFLIB and ENDFIO libraries, and holds the data in the memory as the ENDF object. DeCE works in a following way;
Read entire evaluated nuclear data file, and store all data in the ENDF objects. DeCE assigns one ENDF object to each
section;
Accept user’s command, and manipulate the target object according to the given command and options;
Repeat the operation until quit command; then
Create a new evaluated file.
The structure of DeCE is shown in . In addition to the main DeCE code, the DeCE distribution package includes a collection of short independent programs called the DeCE tools, which do not fit into . Some examples of the DeCE tools are given later.
Figure 3. The schematic illustration of DeCE that uses the ENDFLIB and ENDFIO libraries as an interface between an ENDF-6 file and an ENDF object.
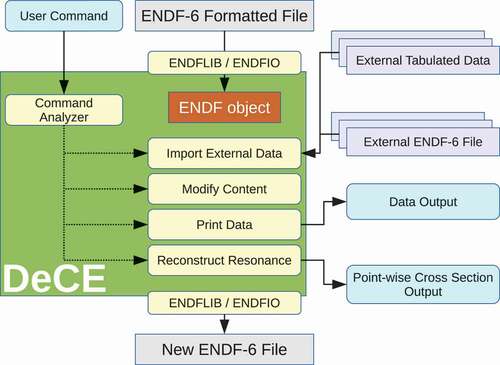
The current version of DeCE recognizes more than 20 commands, which are given in the DeCE manual provided in the DeCE distribution package. Here we present some examples to demonstrate how DeCE works. The DUPLICATE command creates a new object and copy all the source object data into the destination. The DELETE command removes one section, or whole
sections in the same
. The READ command imports external data prepared in a data file into the specified
section. The TABLE command tabulates the stored data in a human-readable format. There are some commands inspired by CRECTJ as well. The CALC command gives the sum, difference, ratio, and product of two sections in
. The MAKE4 command generates the total inelastic scattering cross section from the partial level cross sections.
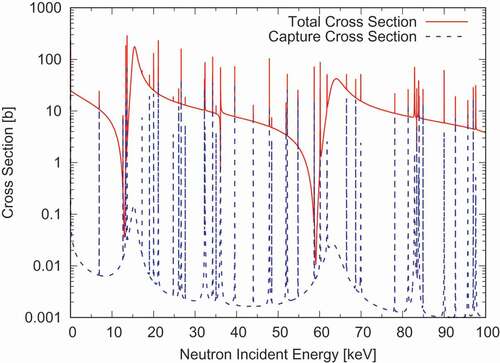
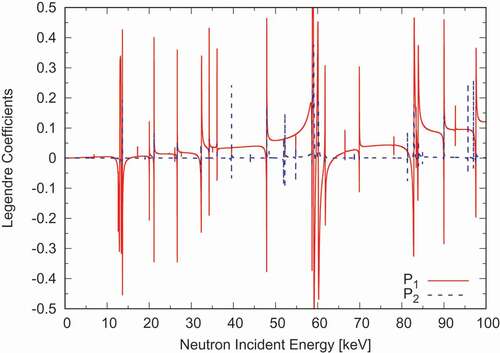
DeCE is also capable of reconstructing point-wise cross-section as well as the elastic scattering angular distributions from the resonance parameters given in . Because DeCE is not a data processing code like FRENDY [Citation11] or NJOY [Citation12], this functionality is somewhat limited. For example, DeCE automatically generates the energy-grid in a relatively simple way that may not guarantee a user-provided tolerance, whereas the processing codes carefully choose the abscissa to satisfy the given tolerance. DeCE does not perform the Doppler broadening either. As examples, reconstructed total and capture cross sections of Ni are shown in . shows the calculated
and
Legendre coefficients [Citation13] in the resolved resonance region. The Reich-Moore resonance parameters are taken from ENDF/B-VIII.
3. Code capabilities and discussions
3.1. The showcase
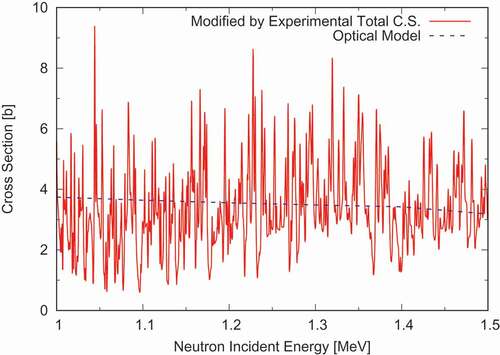
We designed DeCE so flexible that implementation of new commands does not require much work. In fact, since it is an open-source software, the functionality keeps enhancing by incorporating users’ special demands. Here we demonstrate the most frequently used command, CALC, which is similar to the OPERATE command in CRECTJ [Citation7]. Experimental total cross sections in the fast energy range often reveal strong fluctuation even if the energy range is above the resolved resonance region. To evaluate such data, the most realistic method is to trace the experimental total cross section data, as it is impossible to predict the exact fluctuation by nuclear reaction theories. While the elastic scattering cross section, usually calculated by the optical model, shows smooth behavior, as the optical model gives an energy-average cross section by definition. Assuming other reaction channels, neutron radiative capture, fission, inelastic scattering, and so on, vary smoothly, we can reconcile the fluctuating nature in the total cross-section with the optical-model calculated elastic scattering cross section.
First, a non-elastic scattering cross section is reconstructed by summing up all the partial cross sections, then subtract it from the experimental total cross section to evaluate the elastic scattering cross section. The sequence of these operations is like
where the first line adds inelastic scattering and cross sections and put the result into
. The second line instructs DeCE to add the
cross section to
, etc. The re-calculated elastic scattering cross section of
Ni in the 1–1.5 MeV region is compared with the optical model result in , which is above the upper boundary of the resonance range of 812 keV. The fluctuation in the total cross section introduces similar behavior in the elastic scattering channel. Note that the assumption of the smoothness in the other reaction channels is not so correct, as they may fluctuate as well to some extent.
Figure 6. Fluctuation in the elastic scattering cross section introduced by the experimental total cross section of Ni in the 1–1.5 MeV range. DeCE re-distributes the fluctuation on the optical-model calculated smoothed cross section shown by the dashed line.
Since DeCE processes data section-by-section, data access across different sections needs to be handled in a different way. For example, to calculate differential elastic scattering cross sections, we need both the elastic cross section in
and the Legendre coefficients or tabulated distributions given in
. DeCE keeps these data in two objects, and multiplication of
and
is not allowed, because these data could be on the different energy-grids. We provide such capability through the DeCE tools, where ENDFLIB and ENDFIO are still centered. The DeCE tools are a collection of independent short programs provided in the DeCE distribution package. Each of them undertakes a particular task by utilizing ENDFLIB and ENDFIO to read and write ENDF data.
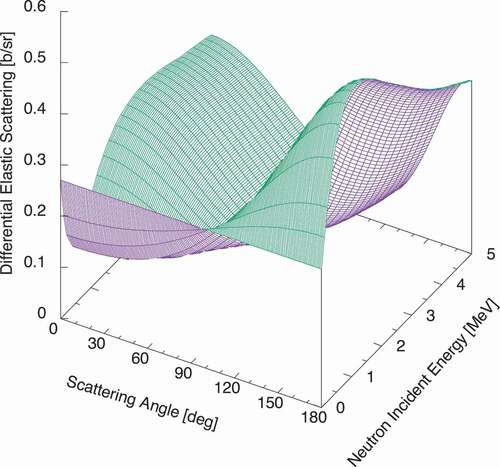
The differential elastic scattering cross section of deuteron is calculated from the angle-integrated cross section and the Legendre coefficients in ENDF/B-VIII, which is shown in . At thermal, the scattering angular distribution is isotropic in the center-of-mass system, and the thermal scattering cross section is 3.395 b, so that the differential cross section is 0.27 b/sr, which one can see in .
Figure 7. Elastic scattering angular distribution of deuteron, reconstructed from the elastic scattering cross section in and the Legendre coefficients in
.
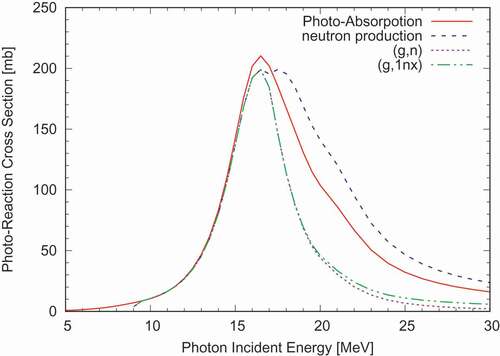
Similarly, we can calculate particle production cross sections, which are usually not given explicitly in the evaluated nuclear data file, and we need to combine and 6. A typical example is the photo-nuclear data library, where the total photon absorption cross section
is given explicitly in
, while
contains production probabilities of particles or residual nuclei. shows the
Nb data in JENDL/PD-2016 [Citation14], processed with one of the DeCE tools. The production cross sections of neutron and charged particles can be calculated easily because they are given in
as ratios to
. The
cross-section, albeit given implicitly, is also calculated unambiguously. However, the experimental
data can be larger than the
cross-section given in the evaluated file, because one-neutron emission concurrent with charged-particle emissions can happen. Therefore, we understand the experimental data to be
. The DeCE tool aggregates all reaction channels which involve one-neutron emission into the
cross-section. This cross section is systematically higher than
above 20 MeV.
3.2. Perspective
As demonstrated, DeCE and its tools help validating evaluated nuclear data files by extracting the data contents in a more convenient form. Obviously, no new physics is involved there, as the data science employed in DeCE just manipulates the evaluated nuclear data only. However, DeCE (or ENDFLIB and ENDFIO) reduces nuclear data users’ unnecessary burden of producing new evaluations, as well as extracting required information from the evaluated nuclear data files. To meet various user-demands that depend on how the nuclear data are utilized, it is important to share the source code, and this is why DeCE is distributed as open-source software.
We envisage that DeCE (or at least ENDFLIB and ENDFIO libraries) will be an indispensable tool for scientists who are involved in the nuclear data production and application fields. There could be some potential subjects in which DeCE can participate in. The simplest case is the nuclear data sensitivity study. As it is very easy to give a small perturbation to the evaluated files with DeCE, one can see how particular nuclear data impact the radiation transport calculation, without modifying neither the processing nor transport simulation code.
ENDFLIB and ENDFIO could be included in a nuclear reaction model code to produce the evaluated data file on-the-fly. In fact, DeCE reads a special file generated by CoH3 [Citation10] and creates a new ENDF-6 file. It is quite possible to produce the evaluated file directly from such nuclear reaction codes without making interim files by incorporating the DeCE libraries (the resonance parameters have to be imported from somewhere). This will create a smooth streamline from the nuclear reaction theory to the Monte Carlo transport simulation, as well as direct feedback from the integral data to the parameters in physics models.
According to this line of thinking, one of the more extreme ideas is to apply DeCE to the resonance analysis. Measured data in the resonance region are often distorted due to the experimental energy resolution, the self-shielding effect, the multiple scattering effect, and so on. To account for all these corrections, one can perform a full Monte Carlo simulation for the entire experimental apparatus to investigate the detector response provided the resonance parameters in the evaluated file. Once sensitivities of the resonance parameters to the detector outputs are calculated, we can perform the least-squares fitting, or more efficient multi-parameter optimization technique, directly to the measured quantities without converting them into the real cross-section data.
In summary, DeCE can be an alternative to CRECTJ, yet DeCE does not emulate all the functionality in CRECTJ. The missing part, such as the group-average, can be done with NJOY. We emphasize that maintaining the user community is indispensable for the development of DeCE as open source. GitHub allows users to report any issues or requests. In fact, we have been improving the source code thanks to the DeCE users, not only internal but also outside LANL. In addition, anybody can modify the source code and request these modifications to be merged into the main branch through the Git mechanism. We always maintain DeCE up-to-date to meet the user’s demand.
4. Conclusion
In this paper, we described ENDF-6 data interface libraries, ENDFLIB and ENDFIO written in C++, which facilitate access to numerical data in the evaluated nuclear data files. The ENDF class defined in ENDFLIB is designed to efficiently store the complicated data structure of ENDF-6. The open-source software DeCE is built on top of these libraries. DeCE processes an ENDF-6 data file interactively to assist in editing the data file, in correcting evaluated data, and in producing a new evaluated file. We demonstrated the capability implemented in the current version of DeCE as well as in the satellite tools by showing some examples; arithmetic operation of two data sections, reconstruction of point-wise cross sections from resonance parameters, production of data sections for particle production cross sections, and production of differential elastic scattering cross sections.
Acknowledgments
The author thanks to P. Talou, M. Paris, M.B. Chadwick, G. Hale, I. Stetcu, and M. White of LANL, I. Thompson of LLNL, D.A. Brown of BNL, N. Otsuka and S. Okumura of IAEA, S. Kunieda of JAEA, and H.I. Kim of KAERI for positive feedback and encouragement to develop DeCE. This work was carried out under the auspices of the National Nuclear Security Administration of the U.S. Department of Energy at Los Alamos National Laboratory under Contract No. 89233218CNA000001.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Shibata K, Iwamoto O, Nakagawa T, et al. JENDL-4.0: a new library for nuclear science and engineering. J Nucl Sci Technol. 2011 Jan;48:1–30.
- Brown DA, Chadwick MB, Capote R, et al. ENDF/B-VIII.0: the 8th major release of the nuclear reaction data library with CIELO-project cross sections, new standards and thermal scattering data. Nucl Data Sheets. 2018;148:1–142.
- Koning AJ, Koning E, Dean J, et al. Status of the JEFF nuclear data library. J Korean Phy Soc. 2011;59:1057–1062.
- Zhuang Y, Liu T, Zhang J, et al. CENDL-3 – Chinese evaluated nuclear data library, version 3. J Nucl Sci Technol. 2002;39(sup 2):37–39.
- Trkov A, Herman M, Brown DA. ENDF-6 formats manual, data formats and procedures for the evaluated nuclear data files, ENDF/B-VI and ENDF/B-VII; 2012. (ENDF-102, BNL-90365-2009 Rev.2).
- Mattoon CM, Beck BR, Patel NR, et al. Generalized nuclear data: a new structure (with supporting infrastructure) for handling nuclear data. Nucl Data Sheets. 2012;113(12):3145–3171.
- Nakagawa T CRECTJ: a computer program for compilation of evaluated nuclear data. Japan Atomic Energy Research Institute; 1999. (JAERI-Data/Code 99-041).
- Iwamoto O. Development of a comprehensive code for nuclear data evaluation, CCONE, and validation using neutron-induced cross sections for uranium isotopes. J Nucl Sci Technol. 2007;44(5):687–697.
- Kunieda S. Status of the R-matrix code AMUR toward a consistent cross-section evaluation and covariance analysis for the light nuclei. EPJ Web Conf. 2017;146:12029.
- Kawano T. CoH3: the coupled-channels and hauser-feshbach code. Proc of CNR2018: International Workshop on Compound Nucleus and Related Topics, LBNL; 2018 Sep 24–28; Berkeley, CA; 2019; J. Escher (Eds.) [to be published].
- Tada K, Nagaya Y, Kunieda S, et al. Development and verification of a new nuclear data processing system FRENDY. J Nucl Sci Technol. 2017;54(7):806–817.
- MacFarlane RE, Kahler AC. Methods for processing ENDF/B-VII with NJOY. Nucl Data Sheets. 2010;111(12):2739–2890.
- Kawano T, Brown DA. Neutron elastic scattering angular distribution in the resolved and unresolved resonance regions. J Nucl Sci Technol. 2015;52(2):274–281.
- Iwamoto N, Kosako K, Murata T. Photonuclear data file. In: Proceedings of the 2015 Symposium on Nuclear Data, JAEA-Conf 2016-004; 2015 Nov 19–20. Tokai-mura, Ibaraki: Ibaraki Quantum Beam Research Center; 2016. p. 53–58.