ABSTRACT
The Pleuroascaceae (Leotiomycetes) is introduced for Phialophora hyalina (section Catenulatae) and its closest relatives based on analyses of DNA sequences of five gene regions and the comparison of cultural and micromorphological characters. The family is resolved as a strongly supported clade that encompasses Pleuroascus and the new anamorph genera Entimomentora and Venustampulla. The latter includes V. parva, a species placed formerly in Scopulariopsis, and V. echinocandica, which is established for the echinocandin-producing isolate BP-5553. Entimomentora includes E. hyalina, a species based on the ex-type strain of Ph. hyalina. Additional isolates identified as Ph. hyalina are distantly related to the Pleuroacaceae and include Psychrophila antarctica (Arachnopezizaceae) and Cryonesomyces dreyfussii, the sole member of the new genus Cryonesomyces (incertae sedis). Isolates identified or deposited as Ph. alba are also not closely related; they include a species for which we propose the name Neobulgaria koningiana (Gelatinodiscaceae) and a second psychrophilic species that we describe as Psychrophila lagodekhiensis. Of the 13 isolates assessed for in vitro antifungal activity, only V. echinocandica inhibited the growth of Candida albicans.
INTRODUCTION
Phialophora was established for anamorphic Ascomycota with one-celled conidia and cylindrical to ampulliform phialides that often terminate as cup-shaped or flaring collarettes at the conidiogenous locus (Medlar Citation1915; Schol-Schwarz Citation1970; Gams and Holubová-Jechová Citation1976; Gams Citation2000). The genus, as monographed by Schol-Schwarz (Citation1970), was a polyphyletic assemblage of poorly defined species that included members of the genera Cadophora and Margarinomyces as well as anamorphs of apothecial and perithecial Ascomycota. Species of Phialophora linked to different sexual states or defined by their ecology were subsequently excluded from the genus (e.g., Catenulifera, Lecythophora, Phaeoacremonium) (Gams and McGinnis Citation1983; Crous et al. Citation1996; Hosoya Citation2002; Harrington and McNew Citation2003; Bogale et al. Citation2010), and phialophora-like anamorphs are now recognized as the members of three classes of ascomycetes (Gams Citation2000; Réblová et al. Citation2017).
Although most species of Phialophora have been linked to sexually reproducing ascomycetes through cultural studies and the comparison of DNA sequences, the phylogenetic disposition of two infrequently collected species with white or lightly pigmented colonies and hyaline conidiogenous cells and conidia remains uncertain. The first of these is Phialophora hyalina, a species with cylindrical phialides and clavate to ellipsoidal conidia that was described in Phialophora section Catenulatae (Gams and Holubová-Jechová Citation1976). This taxon was positioned outside a clade encompassing Phialophora rhodogena (= Hyphodiscus hymeniophilus) and other nonpigmented members of section Catenulatae based on the analysis of partial nuc 28S rDNA sequences, but its position within the Leotiomycetes was not determined (Bogale et al. Citation2010). Phialophora hyalina has since been inferred as the closest relative of BP-5553, a soil isolate from Japan that is the source of the sulfated echinocandin designated as FR190293 (Yue et al. Citation2015). The latter is distinguished by its short, penicillately branched conidiophores, bottle-shaped phialides, and subglobose to ovoidal, hyaline conidia with a truncate base.
The second enigmatic taxon is Ph. alba, a species isolated from the heartwood of beech (van Beyma Thoe Kingma Citation1943) that produces ampulliform phialides with inconspicuous collarettes and subglobose hyaline conidia. Phialophora alba is poorly represented in culture collections, and the name has been applied to few isolates apart from a phialophora-like species from diseased kiwifruit wood in New Zealand (Johnston et al. Citation2010) and a limited number of strains from cold-temperature environments, freshwater sediments, and wood (Ayer and Trifonov Citation1994; Möller and Dreyfuss Citation1996; Ravelet et al. Citation2001; Singh and Singh Citation2012). Isolates of Ph. alba from kiwifruit were connected experimentally to Neobulgaria alba (Leotiomycetes) based on the analyses of nuc rDNA sequences, but their relationship to other members of this class was not resolved (Johnston et al. Citation2010). Examination of additional material deposited as Ph. alba and Ph. hyalina revealed that these isolates could be distinguished from the ex-type strains of both species based on micromorphological and cultural characters.
The present study was motivated by the need for a satisfactory generic and familial classification for isolates identified as Ph. alba and Ph. hyalina based on the comparison of morphological and molecular characters. A second aim of this study was to clarify the nomenclature and phylogenetic positions of fungi shown to be closely related to Ph. alba and Ph. hyalina based on preliminary analyses of rDNA and protein-encoding sequences. Finally, since echinocandins are widely but discontinuously distributed among fungi of the Leotiomycetes (Peláez et al. Citation2011; Yue et al. Citation2015), we were interested in determining whether the isolates most closely related to BP-5553 produced echinocandins and exhibited antifungal activity against Candida albicans.
MATERIALS AND METHODS
Morphological characterization.
—Isolates examined in this study and their sources are listed in . Cultures were maintained on modified Leonian’s agar (MLA) (Malloch Citation1981) at room temperature (20–21 C). For comparative purposes, strains were inoculated in triplicate on filtered oatmeal agar (CBSOA) (Gams et al. Citation1998), MLA, oatmeal agar (OA) (Tuite Citation1969), potato-carrot agar (PCA) (Réblová et al. Citation2017), and potato-dextrose agar (PDA) (Beckton, Dickenson and Co., Sparks, Maryland) in 90-mm Petri dishes. Cultures on CBSOA, MLA, OA, and PCA were incubated at room temperature and examined weekly for up to 28 d. Clearing of the medium around colonies on CBSOA was assessed by holding Petri dishes in front of a light source. Cultures of selected isolates were inoculated on PDA and incubated for 28 d at 4, 10, and 20 C to determine their optimal temperatures of growth. Descriptions of colonies on CBSOA, MLA, OA, and PCA are based on 14- or 28-d-old colonies. Color descriptions of colonies and pigments correspond to the charts of Kornerup and Wanscher (Citation1978). Reverse color is noted only when it differs from that of the colony surface.
Table 1. Isolates examined in this study.
Micromorphological observations were made from isolates grown in slide cultures on MLA following the protocol described by Malloch (Citation1981) or on MLA and PCA. Colonized coverslips and microscope slides were observed in water or Melzer’s reagent using an Olympus BX51 compound microscope (Olympus America, Melville, New York) with differential interference contrast and phase contrast illumination. Images of microscopic structures were captured with an Olympus DP70 camera operated by cellSens Dimension 1.9 imaging software (Olympus America). Measurements of conidia, conidiogenous cells, conidiophores, and hyphae were made from images using the same software. Macroscopic images of colonies were documented using an Olympus C-3030 digital camera with daylight spectrum 5600K 16W light-emitting diode (LED) lights and Dracast diffusion filters (Dracast, San Jose, California). All images were assembled into plates using Adobe Photoshop CS6 (Adobe Systems, San Jose, California).
Antifungal assays and HPLC-DAD–MS analysis.
—Strains evaluated for antifungal activity (indicated in ) were fermented for 14 d using MMK2, MV8, and YES (González-Menéndez et al. Citation2014), supermalt (Ortíz-López et al. Citation2015), and wheat (Li et al. Citation2016) either as static solid or agitated liquid media (220 rpm). Duplicates of each fermentation were extracted by the addition of an equal volume of methyl ethyl ketone (MEK) and acetone followed by shaking for 2 h. The organic phase of the MEK extracts was evaporated with a stream of air, whereas the aqueous phase of the acetone extracts was evaporated under vacuum. Residues were dissolved in dimethyl sulfoxide (DMSO) at 10× the original culture volume, and 20 μL of each DMSO extract was applied to a 4-mm well cut into a plate of yeast malt (YM) agar (González-Menéndez et al. Citation2014) seeded with Candida albicans (ATCC 10231). Plates were incubated at 30 C and examined after 24 h for zones of inhibition against C. albicans. Strain BP-5553 was screened on 11 additional media (see SUPPLEMENTARY FIG. 1) in a zone-of-inhibition assay with C. albicans to verify that it continued to produce the sulfated echinocandin FR190293 and to determine which media yielded strong antifungal activity.
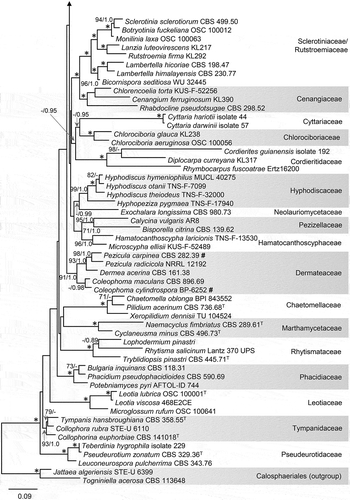
Figure 1. Phylogeny of members of the Leotiomycetes inferred from ML analysis of the concatenated nuc 18S + nuc 28S + mt 18S + RPB2 (four-gene) data set. An asterisk (*) indicates branches with ML BS = 100% and PP values = 1.0. Branch support in nodes ≥70% ML BS and ≥0.90 PP is indicated above or below branches. “T” after an accession number indicates that the strain is an ex-type culture; “#” after an accession number indicates that the strain has demonstrated antifungal activity. Names in bold are taxonomic novelties. Shading is used to differentiate lineages and highlight taxa of interest.
Methods for the extraction, purification, and chromatography of echinocandin analogues followed Li et al. (Citation2016). Chromatographic profiles of crude extract samples were obtained on an Agilent 1260 high-performance liquid chromatograph (Agilent Technologies, Santa Clara, California) equipped with a diode array detector and an Agilent 6120 single quadrupole mass spectrometer. A 10-µL sample of each extract was injected for high-performance liquid chromatography–photodiode array detection mass spectroscopy (HPLC-DAD–MS) analysis on a linear gradient of 10–90% acetonitrile in water (with 0.1% formic acid) for 28 min at a flow rate of 1 mL min−1 through an Agilent Zorbax Eclipse Plus C18 reverse-phase column (4.6 × 150 mm, 5 μm; Agilent Technologies). High-resolution mass spectra were acquired with an Agilent 6520 quadrupole time-of-flight (Q-TOF) system (Agilent Technologies) in the positive ionization mode as described by Li et al. (Citation2016).
DNA extraction, amplification, and sequencing.
—Phylogenetic markers for strains of ATCC 20868, ATCC 42793, ATCC 201333, BP-5553, BP-6252, and NRRL 12192 were extracted from whole genome sequences as described previously (Yue et al. Citation2015; Wingfield et al. Citation2018). Total genomic DNA for strains of CBS 130.74, CBS 177.74, CBS 245.31, CBS 345.73, CBS 120411, CBS 122314, MUCL 9775, and UAMH 918 was extracted from mycelium removed from 14-d-old cultures grown on MLA using the UltraClean Microbial DNA Kit (MoBio Laboratories, Carlsbad, California) following the manufacturer’s protocol for filamentous fungi. Polymerase chain reaction (PCR) amplifications were carried out according to the methods of Réblová et al. (Citation2017).
Primers used for amplifications included (i) ITS1, ITS4, and ITS5 (White et al. Citation1990) for the nuc rDNA internal transcribed spacer ITS1-5.8S-ITS2 region (ITS barcode); (ii) ITS4, NS24, NSSU131, and NSSU97A (White et al. Citation1990; Gargas and Taylor Citation1992; Kauff and Lutzoni Citation2002) for the nuc 18S rRNA gene; (iii) ITS1, ITS5, JS1, JS8, LR0R, LR7, LR8, and NS5 (Vilgalys and Hester Citation1990; White et al. Citation1990; Rehner and Samuels Citation1994; Landvik Citation1996; Vilgalys Citation2018) for the D1, D2, and D3 domains of the nuc 28S rRNA gene; (iv) mrSSU1 and mrSSU3R (Zoller et al. Citation1999) for the mt 18S rRNA gene; and (v) fRPB2-5F and fRPB2-7cR (Liu et al. Citation1999) for segments 5–7 of the second largest subunit of RNA polymerase II (RPB2) gene. PCR products were either purified directly after amplification using UltraClean PCR Clean-up Kit (MoBio Laboratories, Carlsbad, California) or isolated from agarose gel using the High Pure PCR Product Purification Kit (Roche Applied Science, Mannheim, Germany) following the manufacturers’ instructions.
Purified PCR products for ATCC 20868, ATCC 42793, ATCC 201333, BP-5553, BP-6252, and NRRL 12192 were cloned in the pJET1.2/blunt cloning vector (Thermo Fisher Scientific, Waltham, Massachusetts) and sequenced at GENEWIZ (South Plainfield, New Jersey) employing primers included in the CloneJET PCR cloning kit (Thermo Fisher Scientific). Primers used to sequence the PCR products of strains prepared at the Institute of Botany in Průhonice included the following amplification primers and nested primers: (i) ITS1, ITS4, and ITS5 for the ITS; (ii) NS6, NSSU897R, NSSU1088, NSSU1088R, SR7, and SR7R (White et al. Citation1990; Spatafora et al. Citation1995; Kauff and Lutzoni Citation2002; Vilgalys Citation2018) for the nuc 18S; (iii) JS7, LR5, and LR6 (Vilgalys and Hester Citation1990; Landvik Citation1996) for the nuc 28S; (iv) mrSSU2 and mrSSU2R (Zoller et al. Citation1999) for the mt 18S; and (v) RPB2-980F and RPB2-1014R (Reeb et al. Citation2004) for segments 5–7 of the RPB2 gene. Automated sequencing was carried out by GATC Sequencing Service (Cologne, Germany). Raw sequence data were assembled, examined, and manually edited using Genestudio 2.1.1.5 (Genestudio, Suwanee, Georgia) and Sequencher 5.4.1 (Gene Codes, Ann Arbor, Michigan).
Sequence alignment and phylogenetic analyses.
—GenBank accession numbers for sequences generated in this study and previously published sequences of Leotiomycetes and Sordariomycetes (Harrington and McNew Citation2003; Spatafora et al. Citation2007; Baral et al. Citation2009; Schoch et al. Citation2009; Blanchette et al. Citation2010; Bogale et al. Citation2010; Damm et al. Citation2010; Johnston et al. Citation2010; Peterson and Pfister Citation2010; Romero-Olivares et al. Citation2013; Crous et al. Citation2014; Han et al. Citation2014; Taylor et al. Citation2014; Zhang et al. Citation2014; Suija et al. Citation2015; Wang et al. Citation2015; Partel et al. Citation2016) are listed in SUPPLEMENTARY TABLE 1. Homologous ITS sequences of species of Neobulgaria, Psychrophila, and Tetracladium, and members of the Pleuroascaceae, mostly from unidentified isolates and environmental samples, were selected from the top-scoring matches using BLASTn and retrieved from GenBank (NCBI Resource Coordinators Citation2018).
ITS, nuc 18S, nuc 28S, and RPB2 sequences were aligned manually in BioEdit 7.1.8 (Hall Citation1999); the alignment of mt 18S sequences was generated in MAFFT 7 (Katoh and Standley Citation2013) and corrected manually. Single-locus data sets for Leotiomycetes (nuc 18S sequences 77/1796 characters including gaps; nuc 28S 113/1232; mt 18S 50/1993; RPB2 67/1151) and Pleuroascaceae (ITS 19/476; nuc 18S 11/1796; nuc 28S 14/1232; mt 18S 10/1993; RPB2 10/1151) were assessed for conflicts using the 70% reciprocal bootstrap criterion (Mason-Gamer and Kellogg Citation1996) based on the comparison of trees obtained with 1000 bootstrap (BS) replicates with RAxML-HPC 7.0.3 (Stamatakis Citation2006). Conflict-free data sets were concatenated into two multilocus alignments (deposited as TreeBASE 23816) that were subjected to phylogenetic analyses.
Phylogenetic relationships of three isolates of Pleuroascus (CBS 345.73, CBS 537.74, CBS 120411), the ex-type strains of Paecilomyces parvus (CBS 245.31), Phialophora alba (MUCL 9775), Ph. hyalina (CBS 130.74), and Scopulariopsis parvula (UAMH 918), and morphologically similar phialidic hyphomycetes with hyaline conidiophores and vegetative hyphae (ATCC 42793, ATCC 201333, BP-5553, CBS 170.74, CBS 259.65, CBS 122314) were inferred from the analysis of the combined nuc 18S, nuc 28S, mt 18S, and RPB2 sequences of representatives of 32 families recognized in the Leotiomycetes. Based on these results, we performed three subsequent analyses to study the relationships among selected strains. Two Arachnopezizaceae (Arachnopeziza aurata, A. delicatula), two Helotiaceae (Cudoniella clavus, Glarea lozoyensis), and two Sordariomycetes (Jattaea algeriensis, Togniniella acerosa) were used to root individual trees. The first 74 nucleotides (nt) of the nuc 18S and nuc 28S sequences were excluded from the alignment because the majority of these sequences were incomplete at the 5′ end. Introns and ambiguous regions were also excluded from the alignment.
Combined data sets were partitioned into several subsets of nucleotide sites for which we assumed rate heterogeneity. Maximum likelihood (ML) and Bayesian inference (BI) analyses were used to estimate phylogenetic relationships. ML analyses were performed with RAxML-HPC 7.0.3 with a GTRCAT approximation. Nodal support was determined by nonparametric BS analysis with 1000 replicates. BI analyses were performed in a likelihood framework as implemented in MrBayes 3.2.6 (Huelsenbeck and Ronquist Citation2001) through the CIPRES Science Gateway 3.3 (http://www.phylo.org) (Miller et al. Citation2010). For the BI approach, MrModeltest2 2.3 (Nylander Citation2008) was used to infer the appropriate substitution model that best fit the model of DNA evolution. The GTR+I+G model was selected according to the Akaike information criterion for all partitions of data sets for the Leotiomycetes, Neobulgaria, and Pleuroascaceae and the clade encompassing species of Leohumicola, Stamnaria, and Tetracladium, whereas the SYM+G model was selected for the Psychrophila ITS data set. Two Bayesian searches were performed using default parameters. The B-MCMCMC analyses lasted until the average standard deviation of split frequencies was below 0.01 with trees saved every 1000 generations. The first 25% of saved trees, representing the burn-in phase of the analysis, were discarded. The remaining trees were used for calculating posterior probabilities (PPs) of recovered branches. Unless stated, there were no differences in the topologies of trees generated from ML and BI analyses, and all phylogenies are ML trees.
RESULTS
Assay for antifungal activity.
—None of the extracts of cultures of strains of ATCC 42793, ATCC 201333, CBS 130.74, CBS 177.74, CBS 245.31, CBS 259.65, CBS 345.73, CBS 537.74, CBS 120411, CBS 122314, MUCL 9775, and UAMH 918 grown on MMK2, MV8, supermalt, wheat, and YES inhibited the growth of Candida albicans (data not shown). Acetone extracts of BP-5553 grown in most media gave strong activity against C. albicans (SUPPLEMENTARY FIG. 1), and the sulfated echinocandin FR190293 was detected at a retention time of 7.3 min, with the high-resolution mass signal at m/z 1175.54 [M + H+] (SUPPLEMENTARY FIG. 2).
Phylogenetic analysis of the Leotiomycetes.
—The full data set of combined nuc 18S, nuc 28S, mt 18S, and RPB2 sequences of 111 members of the Leotiomycetes consists of 6172 characters and 2978 unique character sites. This class is resolved (ML BS/PP: 100/1.0) in the BI and ML analyses, but the backbone of both trees is statistically unsupported. The BI and ML phylogenies differ in the positions of several families that formed separate, unsupported clades in BI analysis but were grouped in ML analysis. In the ML tree (), the Leotiomycetes includes 26 well-supported clades (≥70/≥0.95) representing 29 families. One strongly supported clade (100/1.0) encompasses the Loramycetaceae, Mollisiaceae, and Vibrisseaceae; a second (100/1.0) includes the Rutstroemiaceae and Sclerotiniaceae. A well-supported lineage (78/0.98) that includes Hyalopeziza leuconica and H. nectrioides is treated as incertae sedis, whereas the Vandijckellaceae/Clade 9 is not supported statistically.
Isolates listed in are inferred as members of six lineages. The echinocandin-producing isolate BP-5533 and strains of Paecilomyces parvus (CBS 245.31, CBS 259.65), Phialophora hyalina (CBS 130.74, CBS 177.74), Pleuroascus nicholsonii (CBS 345.73), Pl. rectipilus (CBS 120411), Pl. rilstonii (CBS 537.74), and Scopulariopsis parvula (UAMH 918) form a strongly supported monophyletic clade (100/1.0) that we describe below as the new family Pleuroascaceae. This family is not related closely to other lineages that include species with cleistothecial ascomata (i.e., Erysiphaceae, Pseudeurotiaceae, and Sclerotiniaceae/Rutstroemiaceae). Three additional echinocandin-producing Leotiomycetes are positioned as follows: the ex-type strain of Glarea lozoyensis (ATCC 20868) belongs to the strongly supported Helotiaceae (86/1.0), whereas Coleophoma cylindrospora (BP-6252) and Pezicula radicicola (NRRL 12192) are inferred as members of a lineage (94/1.0) encompassing other Dermateaceae.
The ex-type strain Phialophora alba (MUCL 9775) is resolved within a well-supported clade (99/1.0) that includes Ascocoryne and Neobulgaria (Gelatinodiscaceae); it is described below as new in the latter genus. Two phialidic hyphomycetes with hyaline mycelia and phialides (ATCC 42793, CBS 122314) group within Psychrophila (100/1.0) in the Arachnopezizaceae (96/1.0); the former isolate is conspecific with Ps. antarctica, whereas the latter is described as a new species in this genus. Finally, a soil isolate from Antarctica (ATCC 201333) is inferred as a member of the clade that also includes Cistella albidolutea, Stamnaria americana, and Vandijckella joannae. This soil isolate, introduced below as a new genus and species, groups with Polyphilus sieberi and “Urceolella” carestiana but without statistical support (<70/0.99).
Phylogenetic analysis of the Pleuroascaceae and topological variation in single- and five-gene phylogenetic trees.
—Relationships within the Pleuroascaceae were examined based on analyses of a data set that included the combined ITS, nuc 18S, nuc 28S, mt 18S, and RPB2 sequences of strains belonging to three genera and the ITS sequences of eight uncultured clones or unidentified isolates retrieved from GenBank. The alignment consisted of 6648 characters and 998 unique character sites. The greatest number of phylogenetically informative characters (unique character sites, RAxML) were provided by the RPB2 (361) and mt 18S (253) sequences as compared with those from the nuc 18S (115), nuc 28S (143), and ITS (126). The trees generated from BI and ML analyses differed only in the position of the sequence of the uncultured fungal clone 3301L16 deposited by Taylor et al. (Citation2014).
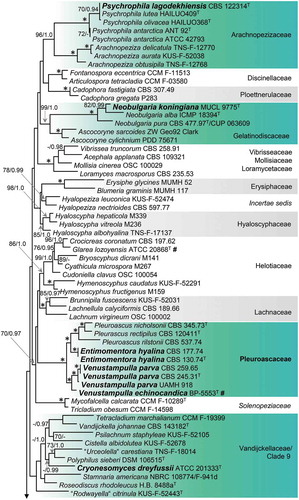
The phylogeny of the Pleuroascaceae based on the analysis of five gene regions () is identical to the clade comprising the species inferred from four loci for 105 species (). Major clades within the family correspond to the genus Pleuroascus (100/1.0), encompassing Pl. nicholsonii, Pl. rectipilus, and Pl. rilstonii, and the newly introduced genera Entimomentora (100/1.0) and Venustampulla (95/1.0).
Figure 2. Phylogeny of the Pleuroascaceae inferred from ML analysis of the concatenated ITS + nuc 18S + nuc 28S + mt 18S + RPB2 (five-gene) data set. Details as in .
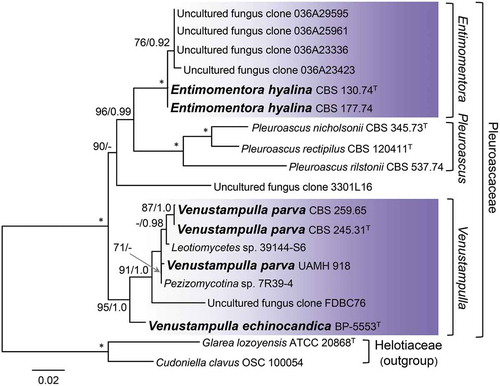
Entimomentora is sister to Pleuroascus and includes E. hyalina, represented by two strains from wheat field soil, and four uncultured clones that may represent an unnamed species. Venustampulla includes isolates identified originally as Cubonia bulbifera (CBS 259.65), Paecilomyces parvus (CBS 245.31), and Scopulariopsis parvula (UAMH 918), for which the combination V. parva is proposed. The genus also includes the echinocandin-producing strain BP-5553 that we describe as V. echinocandica, two unnamed isolates (39144-S6, 7R39-4), and an uncultured clone (FDBC76). The combination of five markers provides strong support (95/1.0) for recognizing Venustampulla as a new genus and BP-5553 as a new species. Reduced statistical support for V. parva in the five-gene analysis likely reflects the lack of sequence data other than the ITS for the two unnamed strains and uncultured clone. Venustampulla echinocandica belongs to a well- to strongly supported subclade representing the genus in single-gene phylogenies inferred from nuc 28S (70% BS) and RPB2 (100% BS) sequences, but it is not retained within Venustampulla in the ITS tree (trees not shown). We did not obtain a readable mt 18S sequence for V. echinocandica.
Phylogenetic analyses of Neobulgaria, Psychrophila, and Cryonesomyces.
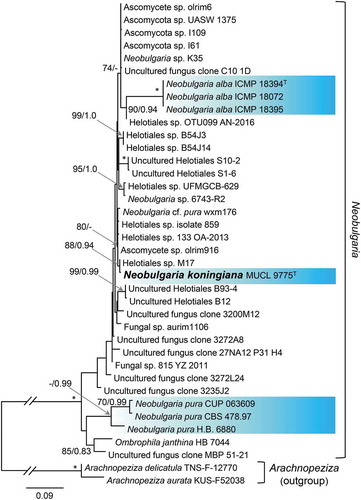
—The relationship of the ex-type strain of Phialophora alba (MUCL 9775) to species of Neobulgaria, represented largely by unidentified strains or environmental samples, was examined based on the analyses of an ITS data set that included 561 characters and 218 unique character sites. Relationships within Neobulgaria are largely unresolved using this marker () except for strains of N. pura (70/0.99), the type species of the genus, N. alba (100/1.0), and a subclade (88/0.94) that includes MUCL 9775 and an unnamed strain (Helotiales sp. M17) isolated from the wood of Alnus glutinosa. This phylogeny and the tree inferred from the five-marker data set () provide strong support for recognizing Ph. alba MUCL 9775 as the new species N. koningiana, which we propose below as a nomen novum.
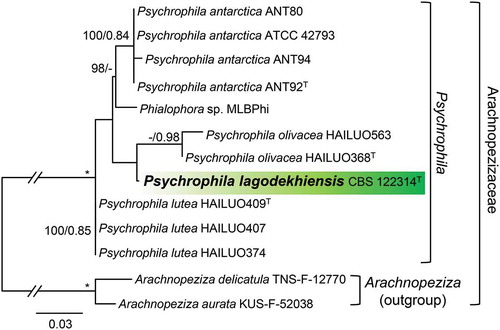
Figure 3. Position of Neobulgaria koningiana (MUCL 9775) within Neobulgaria based on ML analysis of ITS sequences. Details as in .
A second ITS data set consisting of sequences of 11 isolates of Psychrophila includes 510 characters and 112 unique character sites. As illustrated in , Psychrophila is resolved with three subclades representing Ps. antarctica (100/0.84), Ps. lutea (100/0.85), and Ps. olivacea (—/0.98). The strain ATCC 42793, isolated from wood in Sweden, groups within Ps. antarctica. Strain CBS 122314, from soil in the Caucasus Mountains, is sister to Ps. olivacea and is introduced below as a new species, Ps. lagodekhiensis. The soil isolate MLB-Phi (GenBank sequence JN113039, unpublished) from Svalbard is positioned as sister to Ps. antarctica and likely represents a new species.
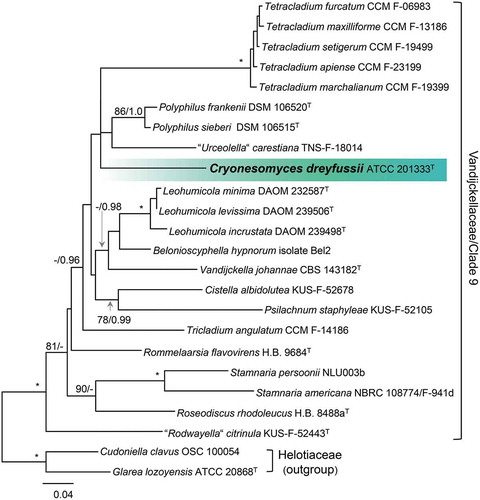
Finally, we explored the relationship of isolate ATCC 201333 to representatives of Clade 9 (Han et al. Citation2014), the Stamnaria lineage (Johnston et al. Citation2019), and Vandijckellaceae (Crous et al. Citation2017) based on comparison of ITS, nuc 18S, nuc 28S, and RPB2 sequences of 22 isolates. Analysis of this alignment, which consists of 4582 characters and 929 unique character sites, resolves this clade as strongly supported (100/1.0). The majority of lineages within this clade lack statistical support except for the subclades represented by Cistella albidolutea and Psilachnum staphyleaea (78/0.99) and species of Tetracladium (100/1.0), Polyphilus (986/1.0), Leohumicola (100/10), and Stamnaria (100/1.0) (). Strain ATCC 201333 belongs to an unsupported subclade that also contains species of Tetracladium, Polyphilus, and “Urceolella” carestiana. Because this strain cannot be placed unequivocally in any known genus based on the comparison of molecular or morphological characters, we introduce it below as a new genus and species, Cryonesomyces dreyfussii.
TAXONOMY
Pleuroascaceae Unter. & Réblová, fam. nov.
MycoBank MB829373
Type genus: Pleuroascus Massee & E.S. Salmon.
Description: Saprobic and coprophilous, lignicolous, or terricolous. Ascomata nonostiolate, spherical, dark, glabrous or setose. Asci prototunicate, globose to subglobose, 8-spored. Ascospores globose to fabiform, 1-celled, hyaline to subhyaline. Conidiogenous cells phialidic, cylindrical to ampulliform, often terminating in a cup-shaped collarette, single on undifferentiated hyphae or grouped on basitonously to penicillately branched conidiophores. Conidia 1-celled, hyaline, subglobose to ellipsoidal or dacryoid, with a truncate base.
Notes: The Pleuroascaceae includes Entimomentora, Venustampulla, and Pleuroascus. The majority of Entimomentora and Venustampulla have been isolated from soil; none are linked to a sexual state. Pleuroascus includes species from dung, decaying wood, and soil that form cleistothecial ascomata, prototunicate asci, and 1-celled hyaline ascospores. Conidial anamorphs are unknown for the members of this genus.
Entimomentora Unter. & Réblová, gen. nov.
MycoBank MB829378
Type species: Entimomentora hyalina (W. Gams) Unter. & Réblová.
Etymology: entimos (Greek), honored, and Mentor, the name of the trusted friend of Odysseus in Homer’s Odyssey and in whose likeness the goddess Athena appears to Odysseus’ son to act as his advisor, a term now used to mean “mentor” or “wise councilor.” This genus is named in honor of the late Walter Gams.
Description: Colonies hyaline to lightly pigmented. Mycelium composed of septate hyaline and darker, monilioid hyphae. Conidiogenous cells phialidic, cylindrical, single and arising from undifferentiated hyphae or from sparingly and basitonously branched conidiophores, each bearing a cylindrical or cup-shaped collarette. Conidia 1-celled, hyaline, ellipsoidal to dacryoid, with a truncate base, slimy. Sexual state not observed.
Entimomentora hyalina (W. Gams) Unter. & Réblová, comb. nov.,
Figure 6. Colonies of species of Entimomentora and Pleuroascus on CBSOA, MLA, OA, and PCA after 28 d. A. E. hyalina (ex-type CBS 130.74). B. Pl. nicholsonii (ex-type CBS 345.73). C. Pl. rectipilus (ex-type CBS 120411). D. Pl. rilstonii (CBS 537.74).
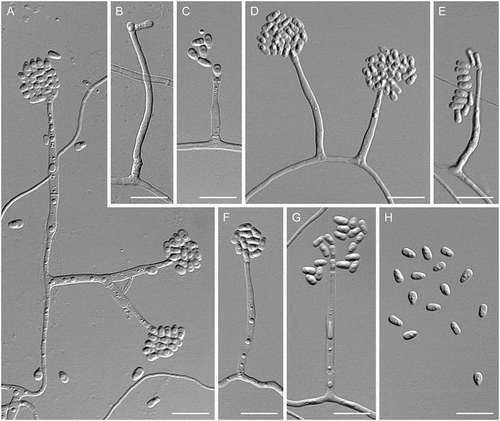
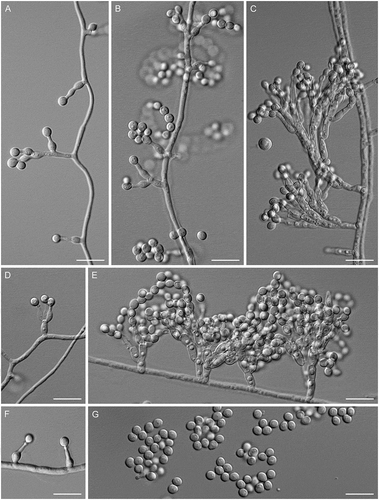
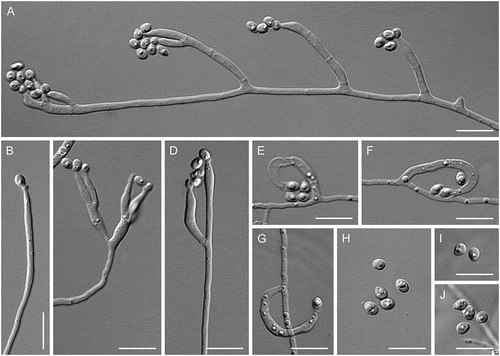
Figure 7. Entimomentora hyalina. A–G. Phialides with conidia (MLA slide culture, 13–17 d). H. Conidia (MLA, 10 d). A–D, H from CBS 177.74; E–G from CBS 130.74. Bars = 10 μm.
MycoBank MB829379
Basionym: Phialophora hyalina W. Gams, Stud Mycol 13:70. 1976.
Description: Hyphae hyaline, septate, smooth-walled, 1.0–2.0 µm wide; thicker-walled, moniliform hyphae up to 14 µm wide produced on CBSOA and OA. Phialides hyaline, single and arising from undifferentiated hyphae or from sparingly and basitonously branched conidiophores, cylindrical, 17–35 × 1.5–3.0 μm (mean ± SD: 27 ± 5.5 × 2.0 ± 0.5 μm), each terminating in a cylindrical or cup-shaped collarette 1.0–4.0 μm deep. Conidia 1-celled, hyaline, ellipsoidal to dacryoid, with a truncate base, 3.0–5.0 × 1.5–2.5 μm (mean ± SD: 4.0 ± 1.0 × 2.0 ± 0.5 μm), forming long chains or accumulating in droplets at the apices of the phialides.
Culture characters: Colonies on CBSOA 51–54 mm diam in 28 d, appearing waxy, flat, aerial mycelium scant, short and restricted to the margin, yellowish-white (4A2). Clearing of the medium to 8 mm from the colony margin observed. Colonies on MLA 30–38 mm diam in 28 d, appearing powdery, aerial mycelium dense and cottony, forming short tufts at the center of the colony, orange-white (5A2), aerial hyphae shorter toward the margin and orange-gray to grayish-orange (5B2–3). Colonies on OA 55–60 mm diam in 28 d, flat to slightly furrowed, otherwise similar in appearance and color to those on MLA. Colonies on PCA 47–50 mm diam in 28 d, similar to those on CBSOA except aerial mycelium denser, particularly at the center, white (5A1) at the center, the remainder of the colony orange-white (5A2). Margin sharply defined and regular on all media.
Specimens examined: GERMANY. Kiel-Kitzeberg, from wheat field soil, 1965, W. Gams. Ex-type culture CBS 130.74; ibid., from wheat field soil, 1964, W. Gams, CBS 177.74.
Notes: Phialophora hyalina was the only member of Phialophora section Catenulatae whose phylogenetic position was unresolved following the recognition of Rhopalophora (Eurotiomycetes) (Réblová et al. Citation2017). This species is morphologically similar to a number of the taxa described in this section (i.e., Ph. brevicollaris, Ph. brachyconia, Ph. rhodogena) that are now recognized as members of the genus Hyphodiscus (Hyphodiscaceae). The ex-type culture of E. hyalina is similar to CBS 177.74 on CBSOA and PCA but forms larger colonies on OA. Colonies of CBS 177.74 on OA are deeply furrowed and covered with tufts of fasciculate aerial hyphae (not shown).
Venustampulla Unter. & Réblová, gen. nov.
MycoBank MB829374
Type species: Venustampulla parva (A.H.S. Br. & G. Sm.) Unter. & Réblová.
Etymology: venustus (Latin), lovely or like Venus, and ampulla (Latin), bottle. In reference to the beautiful, bottle-shaped phialides produced by the members of this genus.
Description: Colonies hyaline to light yellow or orange-brown. Mycelium composed of septate, hyaline or lightly pigmented hyphae. Conidiogenous cells phialidic, cylindrical to ampulliform, lacking a distinct collarette, grouped on short, penicillately branched conidiophores or single and lateral or terminal on undifferentiated hyphae. Conidia 1-celled, hyaline, subglobose to ovoidal, with a truncate base, slimy. Sexual state not observed.
Venustampulla echinocandica Unter., Réblová & Bills, sp. nov.,
Figure 8. Colonies of species of Venustampulla on CBSOA, MLA, OA, and PCA after 28 d. A. V. echinocandica (ex-type BP-5553). B. V. parva (CBS 245.31). C. V. parva (CBS 259.65). D. V. parva (ex-type of Scopulariopsis parvula UAMH 918).
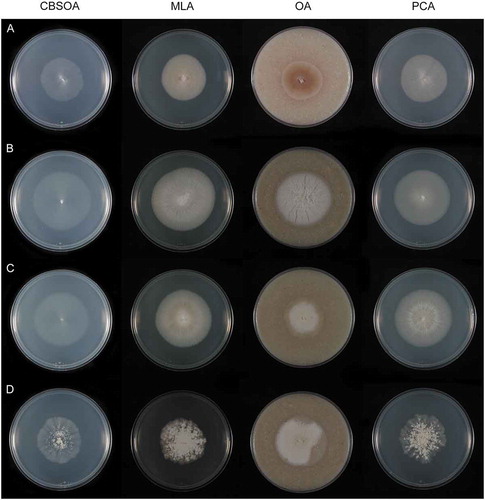
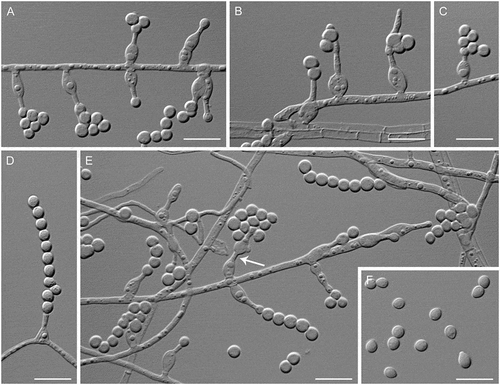
Figure 9. Venustampulla echinocandica (ex-type BP-5553). A–E. Phialides with conidia in chains; arrow indicates a percurrently proliferating phialide (MLA slide cultures, 11 d). F. Conidia (PCA, 12 d). Bars = 10 μm.
MycoBank MB829375
Typification: JAPAN. Kamiina District, from soil (no further collection information available) (holotype NBRC BP-5553, a lyophilized culture permanently preserved in a metabolically inactive state at the NITE Biological Resource Center; isotype UAMH 12038, a dried culture on MLA). Ex-type culture NBRC BP-5553 = Fujisawa No. 16616.
Etymology: In reference to the sulfated echinocandin analogue FR190293 produced by the ex-type strain.
Description: Hyphae hyaline, septate, smooth-walled, and 1.5–2.5 µm wide. Conidiophores absent. Phialides hyaline, sessile, single, lateral or terminal, arising from undifferentiated hyphae, ampulliform, with a swollen base, 5.0–13 × 2.0–5.0 μm (mean ± SD: 8.5 ± 2.0 × 3.0 ± 0.5 μm), tapering abruptly to an elongate, cylindrical tip 1.0–2.0 μm wide, occasionally extending percurrently. Conidia 1-celled, hyaline, subglobose to broadly ellipsoidal, with a truncate base, 2.5–5.0 × 2.5–3.5 μm (mean ± SD: 3.5 ± 0.5 × 3.0 ± 0.5 μm), forming long chains or accumulating in small groups at the apices of the phialides. Chlamydospores, ascomatal initials, and ascomata not observed.
Culture characters: Colonies on CBSOA 37–39 mm diam in 28 d, appearing faintly woolly at the center and mucoid-waxy toward the margin, aerial mycelium scant and restricted to the center of the colonies, white to yellowish-white (4A1–2). Clearing of the medium to 10 mm from the colony margin observed. Colonies on MLA 39–40 mm diam in 28 d, appearing woolly, aerial mycelium short lanose and densest at the center, colony center white to yellowish-white (4A1–2), the remainder of the colony orange-white to pale orange (5A2–3). Colonies on OA 34–38 mm diam in 28 d, appearing powdery, aerial mycelium scant and short, densest at the center, colony center white (A1), becoming brownish-gray to brownish-orange (6C3–6) and then orange-white to orange-gray (5A–B2) toward the margin. Pale orange (6A3) pigment diffusing from the colony margin to 5 mm into the surrounding agar. Colonies on PCA 42–43 mm diam in 28 d, appearing woolly at the center and somewhat felty toward the margin, aerial mycelium sparse and short, densest at the center of the colony, yellowish-white (4A2). Margin sharply defined and entire on all media.
Notes: Venustampulla echinocandica is represented by a single strain (BP-5553 = Fujisawa No. 16616) that produces extracts with antifungal activity against Aspergillus fumigatus and Candida albicans (Kanasaki et al. Citation2006). It is the only member of the Pleuroascaceae thus far known to produce the sulfated echinocandin FR190293. Isolate Fujisawa No. 16616 was identified initially as Tolypocladium parasiticum (= Pochonia parasitica) (Hypocreales), a parasite of rotifers (Barron Citation1980), but its micromorphology is inconsistent with that of Tolypocladium and it has been shown to belong to a clade within the Leotiomycetes that includes Phialophora hyalina (Yue et al. Citation2015). The ex-type isolate has been illustrated as producing erect conidiophores and verticillate phialides (Kanasaki et al. Citation2006; Yue et al. Citation2015), but these structures were not observed on the four media used in our study. The draft genome sequence for BP-5553 (as Phialophora cf. hyalina GenBank assembly ASM335714v1) was published by Wingfield et al. (Citation2018).
This species is distinguished from V. parva by its subglobose conidia and ampulliform phialides that are swollen basally. On OA, this species produces brownish-orange to orange-gray colonies and a pale orange pigment that diffuses 5 mm from the colony margin into the surrounding agar. Older cultures (>10 wk) on MLA are characterized by circular aggregations and rings of thick-walled, melanized hyphae that form in the medium below the colonies (not shown).
Venustampulla parva (A.H.S. Br. & G. Sm.) Unter. & Réblová, comb. nov., ,
Figure 10. Venustampulla parva. A–F. Phialides with conidia (MLA slide cultures, 10–11 d). G. Conidia (MLA, 15 d). A–D, F from CBS 259.65; E, G from ex-type of Paecilomyces parvus CBS 245.31. Bars = 10 μm.
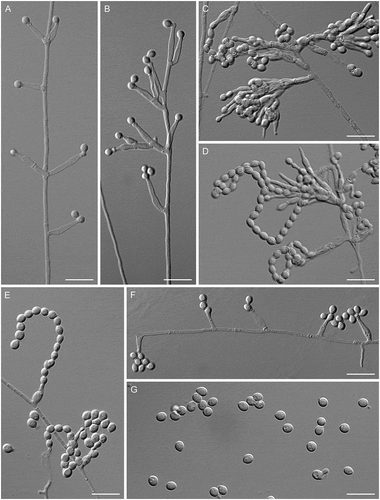
Figure 11. Venustampulla parva (ex-type of Scopulariopsis parvula UAMH 918). A–F. Phialides with conidia (MLA slide cultures, 10–15 d). G. Conidia (MLA, 15 d). Bars = 10 μm.
MycoBank MB829376
Basionym: Paecilomyces parvus A.H.S. Br. & G. Sm., Trans Br Mycol Soc 40:58. 1957.
≡ Scopulariopsis parva (A.H.S. Br. & G. Sm.) Samson, Stud Mycol 6:102. 1974.
= Scopulariopsis parvula F.J. Morton & G. Sm., Mycol Papers 86:65. 1963.
Description: Hyphae hyaline, septate, smooth-walled, and 1.0–2.0 µm wide. Phialides hyaline, single and lateral or terminal on undifferentiated hyphae or grouped on short, penicillately branched conidiophores, cylindrical to ampulliform, 5.5–14.5 × 1.5–3.0 μm (mean ± SD: 8.5 ± 2.0 × 2.0 ± 0.5 μm), tapering gradually to a conidiogenous locus 0.9–1.7 μm wide. Conidia 1-celled, hyaline, broadly ellipsoidal to obovoid, with a truncate base, 3.5–4.5 × 2.5–3.5 μm (mean ± SD: 4.0 ± 0.5 × 3.0 ± 0.5 μm) on MLA and PCA, 3.0–4.5 × 2.0–3.5 μm (mean ± SD: 3.5 ± 0.5 × 2.5 ± 0.5 μm) in slide culture on MLA, forming long chains or accumulating in small groups at the apices of the phialides. Chlamydospores, ascomatal initials, and ascomata not observed.
Culture characters: Colonies on CBSOA 40–52 mm diam in 28 d, appearing powdery at the center and waxy toward the margin, aerial mycelium short, scant and restricted to the center, white to yellowish-white (3A1–2). Colonies on MLA 38–57 mm diam in 28 d, appearing powdery, aerial mycelium short, cottony and forming short fascicles or arranged in overlapping scallops that become less dense toward the margin, yellowish-white to pale yellow (4A2–3), becoming yellowish-gray to grayish-orange (4–5B2–3) toward the margin. Colonies on OA 37–50 mm diam in 28 d, flat to faintly furrowed, appearing powdery-velvety, aerial mycelium cottony, dense and covering most of the colony, occasionally forming short fascicles, yellowish-white (4A2) at the center and becoming white (A1) toward the margin. Colonies on PCA 39–52 mm diam in 28 d, similar to those on MLA but with less dense aerial hyphae, yellowish-white to yellowish-gray (4A–B2), immersed hyphae at margin yellowish-white (4A1). Margin sharply defined and entire on OA; sharply defined to indistinct and entire to undulate on CBSOA, MLA, and PCA.
Specimens examined: Country unknown, from skin infection of a man, A. Stühmer no. 1047. Ex-type culture of Paec. parvus CBS 245.31 = IMI 058410; UAMH 12089, dried culture on MLA. GERMANY. Kiel-Kitzenberg, from wheat field soil, 1963, W. Gams, CBS 259.65. CANADA. ALBERTA: From soil, 1961, J.W. Carmichael. Ex-type culture of Scop. parvula UAMH 918 = ATCC 58635, CBS 209.61, IMI 086943, MUCL 9041; UAMH 12040, dried culture on MLA.
Notes: Paecilomyces parvus was described by Brown and Smith (Citation1957) based on a culture identified as Paec. burci. Samson (Citation1974) transferred Paec. parvus to Scopulariopsis because of its elongating conidiogenous cells and truncate conidia and treated Scop. parvula, a species described as producing annellophores (Morton and Smith Citation1963), as its synonym. Sandoval-Denis et al. (Citation2016) excluded Scop. parva from the Sordariomycetes based on analyses of rDNA sequences of MUCL 9041.
In the phylogeny based on the analysis of five gene regions (), the ex-type strain of Paecilomyces parva (CBS 245.31) grouped with CBS 259.65, an isolate from soil identified originally as Cubonia bulbifera (Leotiomycetes). The nuc 28S sequence of the latter (accessioned in GenBank as FJ176862, Leotiomycetes sp.) was included in the analyses of Bogale et al. (Citation2010) based on its similarity to the nuc 28S sequences of isolates of Ph. hyalina. A third strain of Paec. parva, represented by the ex-type of Scop. parvula (UAMH 918), is resolved as the closest relative of an isolate (7R39-4) from the roof of Shackleton’s hut in the Antarctic identified initially as Solenopeziza solenia (Hyaloscyphaceae) (Blanchette et al. Citation2010).
Isolates of Venustampulla parva cannot be distinguished micromorphologically, but they exhibit a number of differences with respect to colony characteristics. For example, CBS 245.31 produces furrowed colonies on OA, whereas UAMH 918 forms colonies on MLA and PCA that appear scalloped or snowflake-like.
Pleuroascus nicholsonii Massee & E.S. Salmon, Ann Bot 15:330. 1901.
Culture characters: Colonies on CBSOA 37–40 mm diam in 28 d, appearing waxy, flat, aerial mycelium scant, short and restricted to the colony center, immersed mycelium densest at the margin, yellowish-white (4A2). Colonies on MLA 48–50 mm diam in 28 d, appearing woolly, faintly furrowed, aerial mycelium cottony, short, forming low mounds or lumps where dense, yellowish-white to pale yellow (4A2–3) at the center and bearing small, pale yellow droplets of exudate, yellowish-gray to grayish-yellow (4B2–3) at the margin. Reverse yellow to deep yellow (4A6–8). Colonies on OA 50–55 mm diam in 28 d, appearing cottony-velvety, furrowed, resembling those on MLA except aerial mycelium denser, yellowish-white to pastel yellow (3A2–4) at the center and bearing small bright yellow droplets of exudate, becoming white (A1) and then pinkish-white to reddish-gray (7A–B2) at the margin. Brownish-orange (7C6–8) pigment diffusing from the colony margin to 10 mm into the surrounding agar. Reverse grayish-red to brownish-orange (7B–C5–7). Colonies on PCA 40–42 mm diam in 28 d, appearing waxy, aerial mycelium short, cottony, and densest at the colony center, white (A1) at the center, becoming orange-white (5A2) toward the margin. Margin sharply defined and irregular on all media.
Specimen examined: USA. CALIFORNIA: Riverside County, from the dung of pack rat, 1968, R.K. Benjamin. Ex-neotype culture CBS 345.73.
Notes: Pleuroascus nicholsonii was characterized by Malloch and Benny (Citation1973), who described the ex-neotype strain. We did not observe ascomata or the concentric zones of colonies on MLA noted by these workers and suspect that this isolate may have deteriorated with repeated subculturing. We also did not observe conidiogenous cells and conidia on the media employed in our study. Irregularly shaped cells within ascomata were described as conidia by Malloch and Benny (Citation1973) and later interpreted by Plishka et al. (Citation2008) as the remnants of paraphyses.
Pleuroascus nicholsonii is an infrequently encountered species. It is known from only a few additional collections and is found predominantly on the dung of rodents (Plishka et al. Citation2008). Pleuroascus nicholsonii and Pl. rilstonii were first suggested to be closely related to the inoperculate discomycetes by Suh and Blackwell (Citation1999). Developmental evidence supporting the placement of Pl. nicholsonii in the Leotiomycetes was provided by Plishka et al. (Citation2008).
Pleuroascus rectipilus Stchigel, Guarro & Cano, Persoonia 31:287. 2013.
Culture characters: Colonies on CBSOA 20–22 mm diam in 28 d, appearing cottony–faintly powdery, aerial hyphae short, forming small tufts, densest at the center but not uniformly distributed over the colony, light gray (D1) at the colony center, becoming pale gray (B1). Colonies on MLA 29–30 mm diam in 28 d, appearing powdery-velvety, deeply furrowed, aerial mycelium short, dense, and cottony, bluish-gray (22D–E2–3), becoming light gray (B1) with restricted zones of grayish-red to brownish-red (10D5–7), and white (A1) at the margin. Reverse violet-brown (10E4–8) at colony center. Colonies on OA 40–42 mm diam in 28 d, appearing powdery, slightly furrowed, resembling colonies on MLA but with alternating bands of light gray (B1) and light gray to bluish-gray (22C1–2), white (A1) at the margin. Colonies on PCA 25–27 mm diam in 28 d, appearing cottony-powdery at the center and waxy at the margin, aerial mycelium at colony center medium gray (E1) and pale gray (B1) toward the margin, hyphae at margin immersed and brownish-gray (5C2). Margin sharply defined and irregular on all media.
Specimen examined: SPAIN. CANARY ISLANDS: La Laguna, from soil, 1998, A.M. Stchigel. Ex-type culture CBS 120411 = FMR 8954 = MUCL 49873.
Notes: Immature ascomata are evident on all of the media used in our study at 28 d. Mature ascomata, asci, and ascospores conforming to the description provided by Stchigel et al. (Citation2013) are present on PCA after 8 wk. We did not observe conidiogenous cells and conidia on any medium used in our study.
Pleuroascus rectipilus can be distinguished from other members in the genus by its more darkly pigmented (bluish-gray) colonies on MLA and OA that result from the formation of numerous, dark-walled ascomata. The restricted zones of red evident in younger colonies on MLA fade by 28 d. According to Stchigel et al. (Citation2013), Pl. rectipilus differs from Pl. nicholsonii in possessing ascomata with dark, stiff, tuberculate ascomatal hairs.
Pleuroascus rilstonii (C. Booth) Lodha, Taxonomy of fungi (Proc Int Symp Madras, 1973) 1:245. 1978.
≡ Pseudoeurotium rilstonii Booth, Mycol Papers. 83:10. 1961.
≡ Connersia rilstonii (C. Booth) Malloch, Fungi Canadense 32. 1974.
Culture characters: Colonies on CBSOA 20–22 mm diam in 28 d, appearing cottony, aerial mycelium thin, subfloccose, denser toward the colony margin, white (A1). Clearing of the medium to 5 mm from the colony margin observed. Colonies on MLA 38–44 mm diam in 28 d, appearing woolly, aerial hyphae long and covering nearly the entire colony, shorter and appressed at margin, white (A1) and obscuring dark gray (F1) aggregates of ascomata, orange-white to pale orange (6A2–3) at margin. Colonies on OA 45–53 mm diam in 28 d, appearing woolly-felty, resembling colonies on MLA but with alternating bands of white to pale gray (5A–B1) and medium gray (E1), orange-white to pale orange (5A2–3) at the margin. Colonies on PCA 45–54 mm diam in 28 d, appearing cottony, aerial mycelium floccose, covering the entire colony, orange-white (5A2). Margin sharply defined and irregular on MLA and OA, indistinct and undulate on CBSOA and PCA.
Specimen examined: CANADA. ONTARIO: Black Sturgeon Lake, on dead wood of Populus sp., 1973, D.W. Malloch, CBS 537.74.
Notes: Pleuroascus rilstonii is distinguished from the other members of the genus in forming nonsetose ascomata and irregularly ellipsoidal to moon-shaped or broadly allantoid ascospores (Malloch Citation1974; Stchigel et al. Citation2013). In the present study, mature ascomata, asci, and ascospores conforming to the description provided by Malloch (Citation1974) are evident on all media except MLA after 8 wk. The darkly pigmented ascomata are obscured by the dense, cottony or woolly aerial hyphae produced on some media. We did not observe conidiogenous cells and conidia.
Additional Leotiomycetes characterized in this study
Cryonesomyces Unter. & Réblová, gen. nov.
MycoBank MB829380
Type species: Cryonesomyces dreyfussii Unter., Réblová & Bills.
Etymology: cryo (Greek), icy cold, nesos (Greek), island, and mykes (Greek), fungus, in reference to the location where the type species was collected.
Description: Colonies hyaline to lightly pigmented. Mycelium composed of hyaline, septate hyphae. Conidiogenous cells phialidic, cylindrical, single and arising from undifferentiated hyphae or from sparingly and basitonously branched conidiophores, each bearing a cylindrical or cup-shaped collarette. Conidia hyaline, clavate to ellipsoidal and with a truncate base, slimy and accumulating in chains or small groups at the apices of the phialides. Sexual state not observed.
Cryonesomyces dreyfussii Unter., Réblová & Bills, sp. nov.,
Figure 12. Colonies of Neobulgaria and Cryonesomyces on CBSOA, MLA, OA, and PCA. A. Neobulgaria koningiana (ex-type MUCL 9775) after 14 d. B. Cryonesomyces dreyfussii (ex-type ATCC 201333) after 28 d.
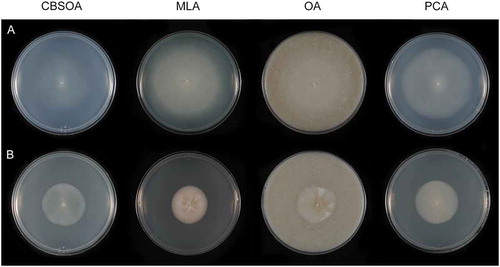
Figure 13. Cryonesomyces dreyfussii (ex-type ATCC 20133). A–F. Phialides with conidia (MLA slide culture, 13 d). G, H. Conidia (MLA slide culture, 13 d). Bars = 10 μm.

MycoBank MB829381
Typification: ANTARCTICA. KING GEORGE ISLAND: From moss, 1991, C. Möller and M.M. Dreyfuss (holotype UAMH 12041, a dried culture on MLA). Ex-type culture ATCC 201333.
Etymology: This species in named for Michael M. Dreyfuss, formerly of the Sandoz natural products laboratory, who organized the collection of the ex-type isolate.
Description: Hyphae hyaline, septate, smooth-walled, 1.5–3.0 µm wide. Phialides hyaline, single and arising from undifferentiated hyphae or from sparingly and basitonously branched conidiophores, cylindrical to ampulliform and tapering toward the apex, 12–19(–22) × 2.5–4.5 μm (mean ± SD: 14.5 ± 2.0 × 3.5 ± 0.5 μm), each terminating in a cylindrical or cup-shaped collarette 1.5–3.0 μm deep and 2.0 × 3.0 μm wide. Conidia hyaline, clavate to ellipsoidal, with a truncate base, 3.5–5.5 × 2.0–3.5 μm (mean ± SD: 4.5 ± 0.5 × 2.5 ± 0.5 μm) on MLA, 2.5–4.5 × 1.5–2.5 μm (mean ± SD: 3.5 ± 0.5 × 2.0 ± 0.5 μm) in slide culture on MLA, forming chains or accumulating in small groups at the apices of the phialides. Chlamydospores, ascomatal initials, and ascomata not observed.
Culture characters: Colonies on CBSOA 38–39 mm diam in 28 d, appearing moist, flat, aerial mycelium absent, white to orange-white (5A1–2). Clearing of the medium to 8 mm from the colony margin observed. Colonies on MLA 30–32 mm diam in 28 d, deeply furrowed at the center, appearing powdery, aerial mycelium short and covering nearly the entire surface of the colony, becoming scant toward the margin, orange-white to grayish-orange (6A3-4). Colonies on OA 33–35 mm diam in 28 d, faintly furrowed, appearing waxy, aerial mycelium sparse and unevenly distributed, colony appearing powdery where aerial hyphae are abundant, yellowish-white to grayish-yellow (4A–B2–3) and becoming white to yellowish-white (4A1–2) toward the margin. Colonies on PCA 38–40 mm diam in 28 d, appearing waxy, aerial hyphae absent, light yellow (4A4–5), becoming white to yellowish-white (4A1–2) toward the margin. Margin sharply defined and regular on all media. Colonies on PDA 32–35 mm diam at 20 C, 30 mm diam at 10 C, and 18–19 mm diam at 4 C in 28 d.
Notes: Cryonesomyces dreyfussii is a psychrotolerant species known from an isolate cultured from moss collected in the Antarctic (Möller and Dreyfuss Citation1996). Colonies on PDA grew fastest at 20 C but were only 2–5 mm smaller in diam at 10 C and more than half the size of the most rapidly growing colonies (i.e., >16 mm diam) at 4 C. This species is reminiscent of Entimomentora hyalina, but it can be distinguished by its shorter, ampulliform phialides, larger conidia, and colonies that are smaller in diam on CBSOA, OA, and PCA.
Neobulgaria koningiana Unter. & Réblová, nom. nov.,
Figure 14. Neobulgaria koningiana (ex-type MUCL 9775). A–G. Phialides with conidia (MLA slide cultures, 10 d). H–J. Conidia (MLA, 10 d). Bars = 10 μm.
MycoBank MB829382
Basionym: Phialophora alba J.F.H. Beyma, Antonie van Leeuwenhoek 9:57. 1943.
Etymology: This species is named for Henriette (Jet) Koning, the collector of the ex-type isolate.
Typification: THE NETHERLANDS. Amersfoort, wood of Fagus sylvatica, H.C. Koning (holotype CBS H-7564; isotypes CBS H-7565, CBS H-7566, CBS H-7567). Ex-type culture CBS 112.43 = IHEM 5885 = MUCL 9775.
Description: Hyphae hyaline, septate, smooth-walled, 1.0–2.5 µm wide. Phialides hyaline, single, arising from undifferentiated hyphae and often curled toward the hyphae on which they are borne, occasionally on basitonously or penicillately branched conidiophores, cylindrical to ampulliform, constricted below the collarette, 9.0–22 × 2.0–3.5 μm (mean ± SD: 15 ± 4.0 × 2.5 ± 0.5 μm). Collarettes indistinct, cup-shaped, 2.0–3.5 μm deep. Conidia hyaline, clavate to ellipsoidal, with a truncate base, 3.5–4.5 × 2.5–3.5 μm (mean ± SD: 4.0 ± 0.5 × 3.0 ± 0.5 μm) on MLA, 3.5–4.5 × 2.0–3.0 μm (mean ± SD: 4.0 ± 0.5 × 2.5 ± 0.5 μm) in slide culture on MLA, slimy and accumulating in small groups at the apices of the phialides. Chlamydospores, ascomatal initials, and ascomata not observed.
Culture characters: Colonies on CBSOA 50–55 mm diam in 14 d, appearing moist to somewhat waxy, flat, lacking aerial hyphae, white (A1). Clearing of the medium to 3 mm from the colony margin observed. Colonies on MLA 75–78 mm diam in 14 d, flat, appearing waxy-mealy or mealy-velvety, slightly furrowed, aerial hyphae sparse especially toward the margin, yellowish-white (4A2). Colonies on OA 50–58 mm diam in 14 d, flat, finely wrinkled at the center, otherwise lacking aerial hyphae, yellowish-white to pale yellow (4A2–3). Colonies on PCA 64–68 mm diam in 14 d, appearing mealy, mycelium entirely immersed and lacking aerial hyphae, white to yellowish-white (4A1–2). Margin indistinct and irregular on all media.
Notes: Neobulgaria koningiana is based on Phialophora alba, but the epithet “alba” is unavailable for this species because of the validly published Neobulgaria alba, the name applied by Johnston et al. (Citation2010) to an apothecial species from kiwifruit vines. Although the kiwifruit pathogen was identified originally as Ph. alba, the ex-type culture of this species (CBS 112.43) was identified as belonging to Paecilomyces based on the comparison of ITS sequences (Johnston et al. Citation2010). The isolate characterized in our study (MUCL 9775) is derived from CBS 112.43 and conforms closely to the descriptions of the Ph. alba provided by van Beyma Thoe Kingma (Citation1943) and Schol-Schwarz (Citation1970). It differs from N. alba in producing smaller, more spherical conidia (3.5–4.5 × 2.5–3.5 μm vs. 4.5–5 × 3–3.5 μm) and phialides that are solitary or grouped on conidiophores with fewer levels of branching. It seems probable, therefore, that the strain representing CBS 112.43 sequenced by Johnston et al. (Citation2010) was a contaminant.
Most of the strains identified as Phialophora alba available from collections originate from soil, healthy and decaying wood, and the decomposing roots; none have been derived from germinating ascospores of Leotiomycetes. Gams (Citation1971) noted the resemblance of Ph. alba to Ph. gregata (= Cadophora gregata), a species responsible for stem rot of legumes (Harrington and McNew Citation2003) that belongs to the Ploettnerulaceae (). Isolates of Ph. alba have been screened for the production of natural products and the ability to degrade pyrene (Ayer and Trifonov Citation1994; Ravelet et al. Citation2001). Singh and Singh (Citation2012) characterized a phialophora-like isolate from water-filled cryoconite holes in Svalbard that they identified as Ph. alba based on its micromorphology and the comparison of nuc 28S sequences. However, the sequence to which this isolate had the highest similarity (99%, AB100618) was obtained from IFM 51363 (Singh and Singh Citation2012), a strain not closely related to any of the species included in our analyses.
Neobulgaria koningiana is the fastest-growing species examined in this study, with colonies that reach the edges of 90 mm Petri plates on all media within 21 d. This species is also distinctive among the isolates we studied in producing curled phialides.
Psychrophila antarctica M.M. Wang & Xing Z. Lui, Persoonia 34:106. 2015. , ,
Figure 15. Colonies of Psychrophila on CBSOA, MLA, OA, and PCA after 28 d. A. Ps. lagodekhiensis (ex-type CBS 122314). B. Ps. antarctica (ATCC 42793).
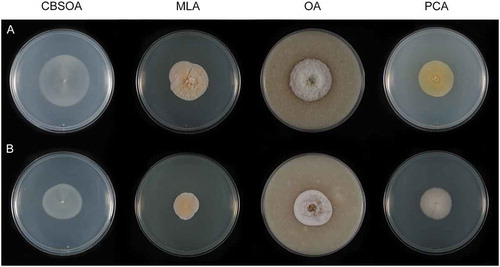
Figure 16. Colonies of Psychrophila on PDA after 28 d at 4, 10, and 20 C. A. Ps. lagodekhiensis (ex-type CBS 122314). B. Ps. antarctica (ATCC 42793).
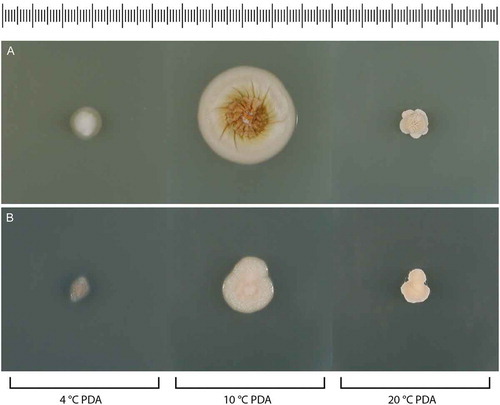
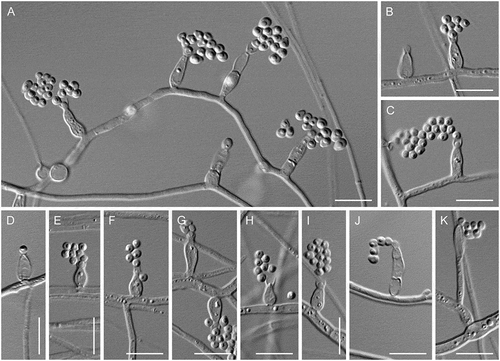
Figure 17. Psychrophila antarctica (ATCC 42793). A–K. Phialides with conidia (MLA slide culture, 13 d). Bars = 10 μm.
Description: Hyphae hyaline, septate, smooth-walled, 1.0–3.0(–4.5) µm wide. Conidiophores absent. Phialides hyaline, single and arising from undifferentiated hyphae, ampulliform, constricted below the collarette, 6.4–11.5(–14.5) × 3.0–5.0 μm (mean ± SD: 9.0 ± 1.5 × 3.5 ± 0.5 μm). Collarettes broadly cup-shaped, 1.5–3.0 μm deep and 2.0–3.0 μm wide. Conidia hyaline, subglobose to dacryoid, with a truncate base, 2.0–3.5 × 1.5–3.0 μm (mean ± SD: 3.0 ± 0.5 × 2.0 ± 0.5 μm), slimy and accumulating in small groups at the apices of the phialides. Chlamydospores, ascomatal initials, and ascomata not observed.
Culture characters: Colonies on CBSOA 29–32 mm diam in 28 d, flat, appearing waxy-mealy, aerial hyphae short and sparse but covering the entire surface of the colony, orange-white (6A2). Faint clearing of the medium to 3 mm from the colony margin observed. Colonies on MLA 20–24 mm diam in 28 d, raised and slightly furrowed, woolly-velvety in appearance, covered in short aerial hyphae, pale orange to orange (6A3–6), white (A1) at the margin. Colonies on OA 29–30 mm diam in 28 d, faintly furrowed, appearing felty-powdery, covered nearly entirely with short aerial that is sparsest at the center, white to orange-white (6A1–2) with a faint brownish-orange (6C4-6) pigment diffusing from the colony margin to 3–5 mm into the surrounding agar. Colonies on PCA 25–29 mm diam in 28 d, appearing powdery, covered entirely in short and dense aerial hyphae, orange-white to pale orange (6A2–3). Margin sharp regular on all media. Colonies on PDA 15–17 mm diam at 10 C and ≤10 mm diam at 4 and 20 C in 28 d.
Specimen examined: SWEDEN. From preservative treated wood, year of origin and collector unknown. ATCC 42793.
Notes: Psychrophila was established by Wang et al. (Citation2015) to accommodate slow-growing, cold-adapted species with unpigmented or brightly colored colonies, pyriform to globose conidia, and phialidic conidiogenous cells. The type species, Ps. antarctica, is known from soil collected in the Antarctic and was characterized as psychrotolerant (= eurypsychrophilic fide Cavicchioli Citation2006), with an optimal growth temperature of 20 C on PDA (Wang et al. Citation2015). The isolate of Ps. antarctica examined in this study was deposited in the ATCC as Ph. hyalina: it conforms closely to the description of the ex-type of Ps. antarctica except that it has an optimal growth temperature of 10 C. Colonies on PDA were not examined for sporulation, but conidiogenous cells and conidia were produced in abundance on CBSOA, MLA, and OA.
Psychrophila lagodekhiensis Unter., Réblová & Bills, sp. nov., ,
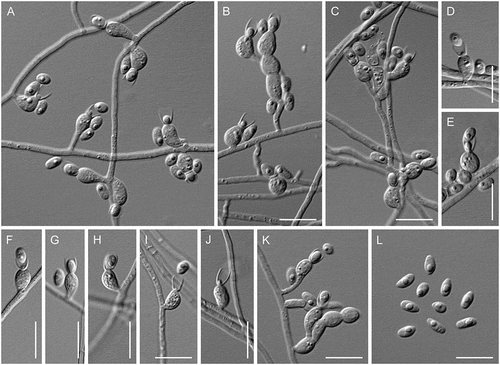
Figure 18. Psychrophila lagodekhiensis (ex-type CBS 122314). A–K. Phialides with conidia (MLA slide culture, 13 d). L. Conidia (MLA slide culture, 13 d). Bars = 10 μm.
MycoBank MB829383
Etymology: This species is named for Lagodekhi, the locality of the ex-type culture.
Typification: GEORGIA. LAGODEKHI: From the rhizosphere of Hedera helix at 645 m, 4 Feb 2007, G.F. Bills (holotype UAMH 12042, a dried culture on MLA). Ex-type culture CBS 122314.
Description: Hyphae hyaline, septate, smooth-walled, 1.4–2.5 µm wide. Phialides hyaline, single and arising from undifferentiated hyphae, occasionally grouped on basitonously branched conidiophores, ampulliform or ventricose, constricted below the collarette, 9.0–13.5 × 3.0–5.0 μm (mean ± SD: 11 ± 1.5 × 4.0 ± 0.5 μm). Collarettes distinct, cup-shaped, 3.5–5.5 μm deep and 2.5–4.0 μm wide. Conidia hyaline, oblong-ellipsoidal to cylindrical, with a truncate base, slimy, 4.0–6.0 × 2.0–3.0 μm (mean ± SD: 5.0 ± 0.5 × 2.5 ± 0.5 μm). Chlamydospores, ascomatal initials, and ascomata not observed.
Culture characters: Colonies on CBSOA 40–45 mm diam in 28 d, flat, appearing moist, mycelium immersed and devoid of aerial hyphae, orange-white (6A2). Clearing of the medium to 5 mm from the colony margin observed. Colonies on MLA 32–34 mm diam in 28 d, flat and mucoid-waxy in appearance in sections devoid of aerial hyphae, otherwise appearing woolly-felty, faintly furrowed and covered in short cottony aerial hyphae that are fasciculate at the center and sparser toward the margin, colony center orange-white to pale orange (6A2–3), pale orange to light orange (6A3-5) toward the margin, and orange-white to pale orange (6A2–3) at the margin and in sectors lacking aerial hyphae. Colonies on OA 35–38 mm diam in 28 d, faintly furrowed, appearing woolly, aerial hyphae long, dense and covering the entire colony, fasciculate at the center, white to light yellow (3A1–5) at the center, white (A1) toward the margin and brownish-orange (6C4-6) at the margin, with a brownish-orange (6C4-6) pigment diffusing from the colony margin to 5 mm into the surrounding agar. Reverse brownish-orange to brown (6C–E3–6). Colonies on PCA 30–32 mm diam in 28 d, appearing waxy, aerial mycelium sparse and short, densest at the center of the colony, grayish-yellow to orange-yellow (4B3–6) at the center becoming light yellow (4A4–5) toward the margin. Margin regular on CBSOA and PCA, irregular on MLA and OA, and sharply defined on all media. Colonies on PDA 28–31 mm diam at 10 C and ≤10 mm diam at 4 and 20 C in 28 d.
Notes: Psychrophila lagodekhiensis is a psychrophilic species known from a single isolate from the rhizosphere of Hedera helix. It is closely related to Ps. olivacea, a soil-inhabiting species from the Hailuogou Glacier that has not been observed to produce philalides and conidia (Wang et al. Citation2015). Among sporulating Psychrophila, Ps. lagodekhiensis can be distinguished by its broad, ampulliform to ventricose phialides and deep, cup-shaped collarettes.
DISCUSSION
The Leotiomycetes comprise morphologically diverse, nonlichenized Ascomycota with apothecial, cleistothecial, or perithecial-like ascomata and asexually reproducing fungi without known relationships to sexual morphs. Their classification, based predominantly on morphologically circumscribed sexual genera and the circumscription of families and orders using genetic markers, varies among authors (Baral Citation2016; Wijayawardene et al. Citation2018; Johnston et al. Citation2019). For example, studies based on the comparison of DNA sequences position the cleistothecial species Pleuroascus nicholsonii and Pl. rilstonii in either the Pseudoeurotiaceae or Helotiales. Pleuroascus, based on Pl. nicholsonii, was described in the Onygenaceae (Eurotiomycetes) by Malloch and Benny (Citation1973), but this species, Pl. rilstonii (as Connersia rilstonii), Leuconeurospora, and Pseudoeurotium were later treated as members of the Pseudoeurotiaceae (Suh and Blackwell Citation1999; Gernandt et al. Citation2001). Pleuroascus nicholsonii and Pl. rilstonii were subsequently shown to be distantly related to Leuconeurospora and Pseudoeurotium (Sogonov et al. Citation2005; Peterson and Pfister Citation2010) and included in the Helotiaceae (Johnston et al. Citation2019).
Our four-gene phylogeny (), based on ribosomal, mitochondrial, and protein-coding loci, resolves Pleuroascus as the member of a strongly supported group (100/1.0) within a lineage (70/0.97) that corresponds to the “helotioid clade” of Johnston et al. (Citation2019). This group, which we recognize as the Pleuroascaceae, encompasses a number of morphologically reduced, phialidic fungi described as species of Entimomentora and Venustampulla. Pleuroascus includes three species with dark-walled, cleistothecial ascomata and one-celled, hyaline or lightly pigmented ascospores (Malloch and Benny Citation1973; Stchigel et al. Citation2013). Conidial anamorphs are not known for the members of this genus (Sogonov et al. Citation2005; Plishka et al. Citation2008; Stchigel et al. Citation2013). Entimomentora and Venustampulla are characterized by their lightly pigmented colonies, conidiogenous cells grouped on penicillately branched conidiophores, and one-celled phialoconidia; none of the members of these new genera have been observed to form ascomata. Entimomentora is based on E. hyalina and is represented by only two isolates. Venustampulla includes V. echinocandica, a species based on the echinocandin-producing isolate BP-5533, and V. parva.
Additional isolates accessioned or identified as Ph. hyalina are distantly related to the Pleuroascaceae. Cryonesomyces dreyfussii, described for a psychrotolerant isolate identified as Ph. hyalina (Möller and Dreyfuss Citation1996), is nested within a lineage encompassing members of Clade 9 sensu Han et al. (Citation2014), the Vandijckellaceae (Crous et al. Citation2017), and the Stamnaria lineage (Johnston et al. Citation2019), as well as Belonioscyphella hypnorum, Tricladium angulatum, and species of Leohumicola. As noted by Johnston et al. (Citation2019), these species appear to share few ecological and morphological characteristics. The Vandijckellaceae/Clade 9 is poorly supported in our four-gene analysis of the Leotiomycetes (), and its structure is not resolved in the five-gene phylogeny (). Within it, Cryonesomyces is inferred as sister to Tetracladium, a monophyletic genus that belongs to a strongly supported clade encompassing other aquatic genera (Mycoarthris and Variocosporium) and species of the ericoid mycorrhizal genus Leohumicola (Kohout et al. Citation2012; Baschien et al. Citation2013). Although phialidic conidiogenesis has not been reported for Tetracladium and its closest relatives, phialoconidial synanamorphs comparable to Cryonesomyces are known for other Leotiomycetes with stauroconidia (Roldán et al. Citation1989; Descals and Moralejo Citation2001; Campbell et al. Citation2006; Seifert et al. Citation2011). Cryonesomyces resembles but is not closely related to Vandijckella joannae, a soil fungus with hyaline, ampulliform conidiogenous cells, conspicuous collarettes, and catenulate, clavate to cylindrical conidia (Crous et al. Citation2017).
A second strain deposited as Ph. hyalina is conspecific with Psychrophila antarctica (Arachnopezizaceae). Isolates described or deposited as Ph. alba are also not closely related. The ex-type culture of Ph. alba is a member of the Gelatinodiscaceae that we recognize as Neobulgaria koningiana; a second isolate is a species of Psychrophila (Arachnopezizaceae) that we describe as Ps. lagodekhiensis. Isolates identified as Phialophora cf. alba that were cultured from moss and plant material collected in the Antarctic (Möller and Dreyfuss Citation1996) could not be located.
The phialidic Leotiomycetes examined in this study share a number of characteristics. First, they are structurally simple and in some cases produce conidiogenous cells that can be challenging to interpret. Second, with the exception of strains now treated as Neobulgara alba, they are collected infrequently. Their paucity in culture collections reflects that they have escaped, or failed to capture, the attention of mycologists. As a consequence, the new species and combinations proposed in this paper are represented by few isolates. Finally, these species belong to families whose members are known from a limited number of cultures (e.g., Pleuroascus, Psychrophila) and as uncultured clones or unnamed isolates (e.g., Entimomentora, Venustampulla). This latter consideration, together with the observation that many isolates originate from undersampled habitats (i.e., cold-temperature environments, rodent dung) suggests that these species are common in some environments.
The Leotiomycetes encompasses the majority of the wild-type strains and species known to produce echinocandins (Peláez et al. Citation2011; Hüttel Citation2017), a group of cyclic hexapeptides that interrupt fungal cell wall synthesis by binding noncompetitively to β-1,3-glucan synthase (Emri et al. Citation2013; Yue et al. Citation2018). Thus, we were hopeful that a phylogenetic- and bioactivity-directed search of additional species related to V. echinocanidica might reveal new echinocandin-producing strains and natural echinocandin variants. However, our search was unsuccessful. Natural echinocandins are of limited use clinically because of their low solubility, limited antifungal spectrum, and tendency for red blood cell lysis (Balkovec et al. Citation2014; Hüttel Citation2017), but semisynthetic derivatives of these metabolites have become first-line treatments of invasive mycoses caused by Aspergillus and Candida (Emri et al. Citation2013; Balkovec et al. Citation2014). The phylogenetic relationships among known echinocandin-producing Leotiomycetes are largely resolved, but the full extent of the diversity of these fungal metabolites, the roles of echinocandins in natural environments, and the mechanisms governing resistance to echinocandins remain poorly understood (Hüttel Citation2017; Yue et al. Citation2018).
We have clarified the taxonomy and phylogenetic relationships of a number of neglected anamorphic Leotiomycetes in public culture collections and anticipate that the descriptions and phylogenies provided in the present study will facilitate the identification of these species, particularly in investigations aimed at measuring the depth of fungal biodiversity, resolving the taxonomy of a number of recognizable but as yet unnamed clades, and the discovery of important natural products. We also hope our work motivates mycologists to collect, culture, and deposit additional isolates of these morphologically reduced fungi, thereby advancing our understanding of these and other phialidic Ascomycota.
Supplemental Material
Download Zip (4.2 MB)ACKOWLEDGMENTS
We are indebted to Dr. Patricia Faasse (Utrecht University) for information concerning the first name of Jet Koning and Dr. Konstanze Bensch (Westerdijk Fungal Biodiversity Institute) for correcting the etymology of Psychrophila lagodekhiensis. Dr. Gary McNeely (Brandon University) and two anonymous reviewers provided constructive comments and suggestions that improved this paper.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s Web site.
Additional information
Funding
LITERATURE CITED
- Ayer WA, Trifonov LS. 1994. Anthraquinones and a 10-hydroxyanthrone from Phialophora alba. Journal of Natural Products 57:317–319.
- Balkovec JM, Hughes DL, Masurekar PS, Sable CA, Schwartz RE, Singh SB. 2014. Discovery and development of first in class antifungal caspofungin (CANCIDAS®)—a case study. Natural Products Reports 31:15–34.
- Baral HO. 2016. Leotiomycetes. In: Jaklitsch W, Baral HO, Lücking R, Lumbsch HT, Frey W, eds. Syllabus of plant families: A. Engler’s syllabus der Pflanzenfamilien 13th ed., part 1/2. Stuttgart, Germany: Borntraeger. p. 157–205.
- Baral HO, De Sloover JR, Huhtinen S, Laukka T, Stenroos S. 2009. An emendation of the genus Hyaloscypha to include Fuscoscypha (Hyaloscyphaceae, Helotiales, Ascomycotina). Karstenia 49:1–17.
- Barron GL. 1980. Fungal parasites of rotifers: a new Tolypocladium with underwater conidiation. Canadian Journal of Botany 58:439–442.
- Baschien C, Tsui CK-M, Gulis V, Szewzyk U, Marvanová L. 2013. The molecular phylogeny of aquatic hyphomycetes with affinity to the Leotiomycetes. Fungal Biology 117:660–672.
- Blanchette RA, Held BW, Arenz BE, Jurgens JA, Baltes NJ, Duncan SM, Farrell RL. 2010. An Antarctic hot spot for fungi at Shackleton’s Historic Hut on Cape Royds. Microbial Ecology 60:29–38.
- Bogale M, Orr MJ, O’Hara MJ, Untereiner WA. 2010. Systematics of Catenulifera (anamorphic Hyaloscyphaceae) with an assessment of the phylogenetic position of Phialophora hyalina. Fungal Biology 114:396–409.
- Brown AHS, Smith G. 1957. The genus Paecilomyces Bainer and its perfect stage Byssochlamys Westling. Transactions of the British Mycological Society 40:17–89.
- Campbell J, Shearer C, Marvanová L. 2006. Evolutionary relationships among aquatic anamorphs and teleomorphs: Lemonniera, Margaritispora, and Goniopila. Mycological Research 110:1025–1033.
- Cavicchioli R. 2006. Cold-adapted archaea. Nature Reviews Microbiology 4:331–343.
- Crous PW, Gams W, Wingfield MJ, van Wyk PS. 1996. Phaeoacremonium gen. nov. associated with wilt and decline diseases of woody hosts anf human infections. Mycologia 88:786–796.
- Crous PW, Quaedvlieg W, Hansen K, Hawksworth DL, Groenwald JZ. 2014. Phacidium and Ceuthospora (Phacidiaceae) are congeneric: taxonomic and nomenclatural implications. IMA Fungus 5:173–193.
- Crous PW, Wingfield MJ, Burgess TI, Carnegie AJ, Hardy GESJ, Smith D, Summerell BA, Cano-Lira JF, Guarro J, Houbraken J, Lombard L, Martín MP, Sandoval-Denis M, Alexandrova AV, Barnes CW, Baseia IG, Bezerra JDP, Guarnaccia V, May TW, Hernández-Restrepo M, Stchigel AM, Miller AN, Ordoñez ME, Abreu VP, Accioly T, Agnello C, Agustin Colmán A, Albuquerque CC, Alfredo DS, Alvarado P, Araújo-Magalhães GR, Arauzo S, Atkinson T, Barili A, Barreto RW, Bezerra JL, Cabral TS, Camello Rodríguez F, Cruz RHSF, Daniëls PP, da Silva BDB, de Almeida DAC, de Carvalho Júnior AA, Decock CA, Delgat L, Denman S, Dimitrov RA, Edwards J, Fedosova AG, Ferreira RJ, Firmino AL, Flores JA, García D, Gené J, Giraldo A, Góis JS, Gomes AAM, Gonçalves CM, Gouliamova DE, Groenewald M, Guéorguiev BV, Guevara-Suarez M, Gusmão LFP, Hosaka K, Hubka V, Huhndorf SM, Jadan M, Jurjević Ž, Kraak B, Kučera V, Kumar TKA, Kušan I, Lacerda SR, Lamlertthon S, Lisboa WS, Loizides M, Luangsa-Ard JJ, Lysková P, Mac Cormack WP, Macedo DM, Machado AR, Malysheva EF, Marinho P, Matočec N, Meijer M, Mešić A, Mongkolsamrit S, Moreira KA, Morozova OV, Nair KU, Nakamura N, Noisripoom W, Olariaga I, Oliveira RJV, Paiva LM, Pawar P, Pereira OL, Peterson SW, Prieto M, Rodríguez-Andrade E, Rojo De Blas C, Roy M, Santos ES, Sharma R, Silva GA, Souza-Motta CM, Takeuchi-Kaneko Y, Tanaka C, Thakur A, Smith MT, Tkalčec Z, Valenzuela-Lopez N, van der Kleij P, Verbeken A, Viana MG, Wang XW, Groenewald JZ. 2017. Fungal Planet description sheets: 625–715. Persoonia 39:270–467.
- Damm U, Fourie PH, Crous PW. 2010. Coniochaeta ( Lecythophora), Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 24:60–80.
- Descals E, Moralejo E. 2001. Water and asexual reproduction in the ingoldian fungi. Botanica Complutensis 24:13–71.
- Emri T, Majoros L, Tóth V, Pócsi I. 2013. Echinocandins: production and applications. Applied Microbiology and Biotechnology 97:3267–3284.
- Gams W. 1971. Cephalosporium-artige Schimmelpilze. Stuttgart, Germany: Gustav Fischer Verlag. 262 p.
- Gams W. 2000. Phialophora and some similar morphologically little-differentiated anamorphs of divergent ascomycetes. Studies in Mycology 45:187–200.
- Gams W, Hoekstra ES, Aptroot A. 1998. CBS course of mycology. 4th ed. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures. 165 p.
- Gams W, Holubová-Jechová V. 1976. Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Studies in Mycology 13:1–99.
- Gams W, McGinnis MR. 1983. Phialemonium, a new anamorph genus intermediate between Phialophora and Acremonium. Mycologia 75:977–987.
- Gargas A, Taylor JW. 1992. Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia 84:589–592.
- Gernandt DS, Platt JL, Stone JK, Spatafora JW, Holst-Jensen A, Hamelin RC, Kohn LM. 2001. Phylogenetics of Helotiales and Rhytismatales based on partial small subunit nuclear ribosomal DNA sequences. Mycologia 93:915–933.
- González-Menéndez V, Asensio F, Moreno C, de Pedro N, Monteiro MC, de la Cruz M, Vicente F, Bills GF, Reyes F, Genilloud O, Tormo JR. 2014. Assessing the effects of adsorptive polymeric resin additions of fungal secondary metabolite chemical diversity. Mycology 5:179–191.
- Hall TA. 1999. BioEdit 5.0.9: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98.
- Han JG, Hosoya T, Sung GH, Shin HD. 2014. Phylogenetic reassessment of Hyaloscyphaceae sensu lato (Helotiales, Leotiomycetes) based on multigene analyses. Fungal Biology 118:150–167.
- Harrington TC, McNew DL. 2003. Phylogenetic analysis places the Phialophora-like anamorph genus Cadophora in the Helotiales. Mycotaxon 87:141–152.
- Hosoya T. 2002. Hyaloscyphaceae in Japan (6): the genus Hyphodiscus in Japan and its anamorph Catenulifera gen. nov. Mycoscience 43:47–57.
- Huelsenbeck JP, Ronquist F. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755.
- Hüttel W. 2017. Structural diversity in echinocandin biosynthesis: the impact of oxidation steps and approaches toward an evolutionary explanation. Zeitschrift für Naturforschung C 72:1–20.
- Johnston PR, Park DC, Manning MA. 2010. Neobulgaria alba sp. nov. and its Phialophora-like anamorph in native forests and kiwifruit orchards in New Zealand. Mycotaxon 113:385–396.
- Johnston PR, Quijada L, Smith CA, Baral HO, Hosoya T, Baschien C, Pärtel K, Zhuang WY, Haelewaters D, Park D, Carl S, López-Giráldez F, Wang Z, Townsend JP. 2019. A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10:1.
- Kanasaki R, Kobayashi M, Fujine K, Sato I, Hashimoto M, Takase S, Tsurumi Y, Fujie A, Hino M, Hashimoto S, Hori Y. 2006. FR227673 and FR190293, novel antifungal lipopeptides from Chalara sp. no. 22210 and Tolypocladium parasiticum no. 16616. The Journal of Antibiotics (Tokyo) 59:158–167.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30:772–780.
- Kauff F, Lutzoni F. 2002. Phylogeny of the Gyalectales and Ostropales (Ascomycota, Fungi): among and within order relationships based on nuclear ribosomal RNA small and large subunits. Molecular Phylogenetics and Evolution 25:138–156.
- Kohout P, Sýkorová Z, Čtvrtlíková M, Rydlová J, Suda J, Vohník M, Sudová R. 2012. Surprising spectra of root-associated fungi in submerged aquatic plants. FEMS Microbiology Ecology 80:216–235.
- Kornerup A, Wanscher JH. 1978. Methuen handbook of colour, 3rd ed. London: Eyre Methuen. 252 p.
- Landvik S. 1996. Neolecta, a fruit-body-producing genus of the basal ascomycetes, as shown by SSU and LSU rDNA sequences. Mycological Research 100:199–202.
- Li Y, Yue Q, Krausert NM, An Z, Gloer JB, Bills GF. 2016. Emestrins: anti-Cryptococcus epipolythiodioxopiperazines from Podospora australis. Journal of Natural Products 79:2357–2363.
- Liu YJ, Whelen S, Hall BD. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16:1799–1808.
- Malloch D. 1974. Connersia rilstonii. Fungi Canadenses No. 32. Ottawa, Canada: National Mycological Herbarium, Agriculture Canada. 2 p.
- Malloch D. 1981. Moulds: their isolation, cultivation and identification. Toronto, Canada: University of Toronto Press. 97 p.
- Malloch D, Benny GL. 1973. California ascomycetes; four new species and a new record. Mycologia 65:648–660.
- Mason-Gamer RJ, Kellogg EA. 1996. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Systematic Biology 45:524–545.
- Medlar J. 1915. A new fungus, Phialophora verrucosa, pathogenic for man. Mycologia 7:200–203.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, Louisiana, USA. p. 45–52.
- Möller C, Dreyfuss MM. 1996. Microfungi from Antarctic lichens, mosses and vascular plants. Mycologia 88:922–993.
- Morton FJ, Smith G. 1963. The genera Scopulariopsis, Microascus and Doratomyces. Mycological Papers 86:1–96.
- NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 46:D8–D13.
- Nylander J. 2008. MrModeltest2 v. 2.3 (Program for selecting DNA substitution models using PAUP*). Uppsala, Sweden: Evolutionary Biology Centre.
- Ortíz-López FJ, Monteiro MC, González-Menéndez V, Tormo JR, Genilloud O, Bills GF, Vincente F, Zhang C, Roemer T, Singh, Reyes F. 2015. Cyclic colisporifungin and linear cavinafungins, antifungal lipopeptides isolated from Colispora cavincola. Journal of Natural Products 78:468–475.
- Partel K, Baral HO, Tamm H, Põldmaa K. 2016. Evidence for the polyphyly of Encoelia and Encoelioideae with reconsideration of respective families in Leotiomycetes. Fungal Diversity 82:183–219.
- Peláez F, Collado J, Platas G, Overy DP, Martín J, Vicente F, González de Val A, Basilio A, de la Cruz M, Tormo JR, Fillola A, Arenal F, Villareal M, Rubio V, Baral HO, Galán R, Bills GF. 2011. The phylogeny and intercontinental distribution of the pneumocandin-producing anamorphic fungus Glarea lozoyensis. Mycology 2:1–17.
- Peterson KR, Pfister DH. 2010. Phylogeny of Cyttaria inferred from nuclear and mitochondrial sequence and morphological data. Mycologia 102:1398–1416.
- Plishka MJR, Tsuneda A, Currah RS. 2008. Evidence of apothecial ancestry in the cleistothecial ascomata of Pleuroascus nicholsonii. Mycological Research 112:1319–1326.
- Ravelet C, Grosset C, Krivobok S, Montuella B, Alary J. 2001. Pyrene degradation by two fungi in a freshwater sediment and evaluation of fungal biomass by ergosterol content. Applied Microbiology and Biotechnology 56:803–808.
- Réblová M, Untereiner WA, Štěpánek V, Gams W. 2017. Disentangling Phialophora section Catenulatae: disposition of taxa with pigmented conidiophores and recognition of a new subclass, Sclerococcomycetidae (Eurotiomycetes). Mycological Progress 16:27–46.
- Reeb V, Lutzoni F, Roux C. 2004. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Molecular Phylogenetics and Evolution 32:1036–1060.
- Rehner SA, Samuels GJ. 1994. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research 98:625–634.
- Roldán A, Descals E, Honrubia M. 1989. Pure culture studies on Tetracladium. Mycological Research 93:452–464.
- Romero-Olivares AL, Baptista Rosas RC, Escalante AE, Bullock SH, Riquelme M. 2013. Distribution patterns of Dikarya in arid and semiarid soils of Baja California, Mexico. Fungal Ecology 6:92–101.
- Samson RA. 1974. Paecilomyces and some allied hyphomycetes. Studies in Mycology 6:1–119.
- Sandoval-Denis M, Gené J, Sutton DA, Cano-Lira JF, de Hoog GS, Decock CA, Wiederhold NP, Guarro J. 2016. Refining Microascus, Scopulariopsis and allied genera. Persoonia 36:1–36.
- Schoch CL, Sung GH, López-Giráldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, de Hoog GS, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh SO, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lücking R, Büdel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O’Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW. 2009. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Systematic Biology 58:224–239.
- Schol-Schwarz MB. 1970. Revision of the genus Phialophora (Moniliales). Persoonia 6:59–64.
- Seifert K, Morgan-Jones G, Gams W, Kendrick B. 2011. The genera of hyphomycetes. Utrecht, The Netherlands: CBS KNAW Fungal Biodiversity Centre. 997 p.
- Singh P, Singh SM. 2012. Characterization of yeast and filamentous fungi isolated from cryoconite holes in Svalbard, Arctic. Polar Biology 35:575–583.
- Sogonov MV, Schroers H-L, Gams W, Dijksterhuis J, Summerbell RC. 2005. The hyphomycetes Teberdinia hygrophila gen. nov., sp. nov. and related anamorphs of Pseudeurotium species. Mycologia 97:695–709.
- Spatafora JW, Mitchell TG, Vilgalys R. 1995. Analysis of genes coding for small-subunit rRNA sequences in studying phylogenetics of dematiaceous fungal pathogens. Journal of Clinicical Microbiology 33:1322–1326.
- Spatafora JW, Sung GH, Johnson D, Hesse C, O’Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, Reeb V, Gueidan C, Fraker E, Lumbsch T, Lücking R, Schmitt I, Hosaka K, Aptroot A, Roux C, Miller AN, Geiser DM, Hafellner J, Hestmark G, Arnold AE, Büdel B, Rauhut A, Hewitt D, Untereiner WA, Cole MS, Scheidegger C, Schultz M, Sipman H, Schoch CL. 2007. A five-gene phylogenetic analysis of the Pezizomycotina. Mycologia 98:1020–1030.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690.
- Stchigel AM, Guarro J, Cano-Lira JF. 2013. Fungal Planet 210—Pleuroascus rectipilus. Persoonia 31:286–287.
- Suh SO, Blackwell M. 1999. Molecular phylogeny of the cleistothecial fungi placed in Cephalothecaceae and Pseudeurotiaceae. Mycologia 91:836–848.
- Suija A, Ertz D, Lawrey D, Diederich P. 2015. Multiple origin of the lichenicolous life habit in Helotiales, based on nuclear ribosomal sequences. Fungal Diversity 70:55–72.
- Taylor DL, Hollingsworth TN, McFarland JW, Lennon NJ, Nusbaum C, Ruess RW. 2014. A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecological Monographs 84:3–20.
- Tuite J. 1969. Plant pathological methods. Minneapolis, Minnesota: Burgess Publishing Co. 239 p.
- Van Beyma Thoe Kingma JFH. 1943. Beschreibung der im Centraalbureau voor Schimmelcultures vorhandenen Arten der Gattungen Phialophora Thaxter und Margarinomyces Laxa, nebst Schlüssel zu ihrer Bestimmung. Antonie van Leeuwenhoek 9:51–76.
- Vilgalys R. 2018. Conserved primer sequences for PCR amplification of fungal rDNA. [cited 2019 Aug 30]. Available from: https://sites.duke.edu/vilgalyslab/rdna_primers_for_fungi/
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172:4238–4246.
- Wang M, Jiang X, Wu W, Hao Y, Su Y, Cai L, Xiang M, Liu X. 2015. Psychrophilic fungi from the world’s roof. Persoonia 34:100–112.
- White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols: a guide to methods and applications. San Diego, California: Academic Press. p. 315–322.
- Wijayawardene N, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. 2018. Outline of Ascomycota: 2017. Fungal Diversity 88:167–263.
- Wingfield BD, Bills GF, Dong Y, Huang W, Nel WJ, Swalarsk-Parry BS, Vaghefi N, Wilken PM, An Z, de Beer ZW, De Vos L, Chen L, Duong TA, Gao Y, Hammerbacher A, Kikkert JR, Li Y, Li H, Li K, Li Q, Liu X, Ma X, Naidoo K, Pethybridge SJ, Sun J, Steenkamp ET, van der Nest MA, van Wyk S, Wingfield MJ, Xiong C, Yue Q, Zhang X. 2018. Draft genome sequence of Annulohypoxylon stygium, Aspergillus mulundensis, Berkeleyomyces basicola (syn. Thielaviopsis basicola), Ceratocystis smalleyi, two Cercospora beticola strains, Coleophoma cylindrospora, Fusarium fracticaudum, Phialophora cf. hyalina, and Morchella septimelata. IMA Fungus 9:185–208.
- Yue Q, Chen L, Zhang X, Li K, Sun J, Liu X, Bills GF. 2015. Evolution of chemical diversity in the echinocandin lipopeptide antifungal metabolites. Eukaryotic Cell 14:698–718.
- Yue Q, Li Y, Chen L, Zhang X, Liu X, An Z, Bills GF. 2018. Genomics-driven discovery of a novel self–resistance mechanism in the echinocandin-producing fungus Pezicula radicicola. Environmental Microbiology 20:3154–3167.
- Zhang T, Victor TR, Rajkumar SS, Li X, Okoniewski JC, Hicks AC, Davis AD, Broussard K, LaDeau SL, Chaturvedi S, Chaturvedi V. 2014. Mycobiome of the bat white nose syndrome affected caves and mines reveals diversity of fungi and local adaptation by the fungal pathogen Pseudogymnoascus (Geomyces) destructans. PLoS ONE 9:e108714.
- Zoller S, Scheidegger C, Sperisen C. 1999. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31:511–516.