ABSTRACT
Piptoporellus baudonii is proposed as a new combination for Laetiporus baudonii in the Polyporales (Basidiomycota) based on morphological and molecular features. This parasitic macrofungus attacks cashew trees, Eucalyptus, cassava, Tectona, and some indigenous trees in southern regions of Tanzania and poses a serious threat to agroforestry and livelihood conditions in the area. Phylogenetic trees were produced from partial sequences of three rDNA gene regions and a portion of translation elongation factor 1-alpha (TEF1) gene of Laetiporus baudonii for comparisons with samples from the antrodia clade. Our results reveal a strongly supported group of L. baudonii with Piptoporellus in Fomitopsidaceae. Piptoporellus baudonii shares many morphological features with other members of Piptoporellus but differs from them in having broadly ellipsoid or rarely ovoid basidiospores. Both morphological and phylogenetic evidence justify the placement of L. baudonii in Piptoporellus together with the three other known species in the genus.
INTRODUCTION
Laetiporus baudonii (Pat.) Ryvarden (Ryvarden Citation1991) is a large parasitic polypore fungus well known in Africa. Reports suggest that the species afflicts a wide range of economically important plants in various parts of Africa (Van der Westhuizen Citation1973), including Congo (Patouillard Citation1914), on Manihot in Madagascar (Heim Citation1931), on Cassia siamea, Khaya senegalensis, and Citrus species in Ghana (Ofosu-Asiedu Citation1975), in tea plantations in Malawi (Rattan and Pawsey Citation1981), on Eucalyptus in South Africa (Van der Westhuizen Citation1973), on Acacia tortilis in Kenya, on Anacardium occidentale (Cashew tree) in Tanzania, and on various indigenous plants (Sijaona Citation2007). Death of the host plants is associated with the appearance of large bright orange-yellow poroid basidomata at the bases of trunks and on the ground among the infected trees. These fruit bodies develop from a pseudosclerotial tissue composed of a mat of coarse creamy-yellowish mycelium and sand grains that cover the roots and underground parts of the stems of afflicted trees (Van der Westhuizen Citation1973). Root infection causes early symptoms of loss of the deep green color of leaves and the yellowing of leaves on individual branches, followed by frequent and rapid wilting of the leaves (Rattan and Pawsey Citation1981).
Laetiporus baudonii belongs to the order Polyporales (Basidiomycota), which includes more than 1800 species (Kirk et al. Citation2008). Most species in this order occur on wood. Seven major clades have been recognized in the Polyporales—the core polyporoid, residual polyporoid, phlebioid, gelatoporia, tyromyces, fragiliporia, and antrodia clades (Binder et al. Citation2005; Zhao et al. Citation2015; Justo et al. Citation2017). Justo et al. (Citation2017) assigned family names to 18 clades in their formal classification. The antrodia clade encompasses many genera, including Laetiporus Murrill and Phaeolus (Pat.) Pat. in Laetiporaceae Jülich and Piptoporus P. Karst. and Fomitopsis P. Karst. in Fomitopsidaceae Jülich.
Since it was first described from Congo, Polyporus baudonii Pat. (Patouillard Citation1914) has been less well understood in a systematic framework that has resulted in the naming of several taxonomic synonyms (Westerhuizen Citation1973). In a study investigating the morphology of fruit bodies and cultures isolated from a parasitic fungus found in South Africa, and comparison with the descriptions of the type specimens of Polyporus baudonii and Phaeolus manihotis R. Heim, Van der Westhuizen (Citation1973) observed morphological similarities between the studied parasitic fungus and Phaeolus manihotis, with the former generally having larger basidiomata than the latter. Van der Westhuizen (Citation1973) confirmed the morphological resemblance between Phaeolus manihotis and Polyporus baudonii as suggested by Browne (Citation1968). The study also referred to the fungus as Polyporus baudonii due to the observation of a dimitic hyphal system that differed from that of the type species of Phaeolus (Donk Citation1960), which has a monomitic hyphal system. Furthermore, Van der Westhuizen (Citation1973) observed complex morphological features that did not match those of any known genus at that time. Because of this, Ryvarden described the new genus Pseudophaeolus Ryvarden, with Ps. baudonii (Pat.) Ryvarden as the type (Ofosu-Asiedu Citation1975). Later, Ryvarden (Citation1991) accepted P. baudonii in Laetiporus as L. baudonii (Pat.) Ryvarden.
The association of a polyporoid macrofungus with wilting of cashew nut trees, cassava, and Eucalyptus trees in the Mtwara and Lindi regions of southern Tanzania has been reported (Sijaona Citation2003, Citation2007). Morphologically, this fungus resembled L. baudonii. In order to ascertain its identity, and given the inconsistencies in classifications based on morphological features, we incorporated molecular data for an accurate placement. This study aims to ascertain the phylogenetic position of L. baudonii using a four-gene region data set based on analyses of partial sequences from three nuc rDNA gene regions and translation elongation factor 1-alpha (TEF1).
MATERIALS AND METHODS
Study site.—
The material studied originated from the Mtwara region in southern Tanzania (ca. 10°10′–11°30′S to 37°58′–40°25E), where cashew is a major cash crop that is frequently affected by this fungus. The region is mostly characterized by a high relative humidity (87% in Feb–Jun, which is usually the long rainy season), whereas the lowest relative humidity of 63% occurs in Sep–Oct. Temperatures vary with cold months (Jun–Sep) with an average of 19.5 C, whereas hot months reach above 30 C (Sep–Dec). The soils are sandy loam or red clay soil, receiving an annual rainfall of 900–1200 mm (Sijaona Citation2003; Sijaona and Shija Citation2005).
Field sampling.—
An unidentified fungus was first observed by the staff at the Naliendele Agricultural Research Institute (NARI). A field trip was arranged by the first author during the rainy season of 2011. Basidiomata were collected, and each collection locality was recorded using Global Positioning System (GPS) (Tibuhwa et al. Citation2010). Prior to collecting, mushrooms were photographed in situ (–D). Field identification features such as sporocarp shape, color, smell, and color changing on bruising were recorded. Collected samples were brought to the Department of Molecular Biology and Biotechnology Laboratory at the University of Dar es Salaam (UDSM) where parts of the mushrooms were oven dried at 50 C for 8 h. Vouchers were deposited at UDSM and UPS herbaria, the latter registered in Index Herbariorum (Thiers [continuously updated]). Color descriptions were based on Kornerup and Wanscher (Citation1962).
Figure 1. Piptoporellus baudonii in situ. A. Basidioma occurring on Eucalyptus (Tibuhwa 1098.2014). B. Turtle eating an old fruiting body. C. Basidioma attached to the root of Eucalyptus (Tibuhwa 1098.2014). D. Dying Eucalyptus with basidioma (Tibuhwa 1098.2014) forming at the base of the tree

Four collections of Pseudophaeolus baudonii (Pat.) Ryvarden (one from Zimbabwe, one from Senegal, one from Ghana, and one from Uganda) were obtained from the herbarium at Oslo University.
Microscopy.—
Microscopic characters were recorded from specimens preserved by dehydration using silica gel and later observed in a 10% ammonium solution in an aqueous solution of Congo red. Twenty measurements of basidiospores and basidia were analyzed statistically, with the results presented as (min)A−SD–A+SD(max), where min is smallest observed value, A is the arithmetic average, SD is the standard deviation, and max is largest observed value for the measured specimen. Basidiospore characters, which include their size, shape, and reactions to Melzer’s reagent, were observed. The hyphal system (monomitic, dimitic, or trimitic) was studied as well as the type of septa (simple septa or clamped septa in generative hyphae).
Molecular study.—
This part of the study was carried out at the Department of Organismal Biology, Uppsala University. Total DNA was extracted from the inner part of the basidiomata, preferably from the hymenium to avoid contamination, following the protocol of the Plant Genomic DNA Extraction Kit (Qiagen, Hilden, Germany). Diluted (10−1–10−3) and undiluted DNA was used for polymerase chain reaction (PCR) amplifications. For herbarium material, nested PCR was employed due to low yields from initial PCR. The 5′ end of the nuc 28S rDNA (28S), nuc rDNA internal transcribed spacer region ITS1-5.8S-ITS2 (ITS), a portion of nuc 18S rDNA (18S), and a portion of TEF1 were amplified. Primers used for amplification and sequencing included LR0R, LR5, and LR7 (Vilgalys and Hester Citation1990) for 28S; ITS1F (Gardes and Bruns Citation1993), 5.8S (Vilgalys and Hester Citation1990), ITS3, ITS4, and ITS5 (White et al. Citation1990) for ITS; NS1, NS7, and NS4 for 18S (White et al. Citation1990); and EF1-983F, EF-2218R, and EF1-1567R for TEF1 (Rehner and Buckley Citation2005). For PCR amplification, we used the AccuPower PCR PreMix (Bioneer, Daejeon, Korea), adding 3 µL diluted or undiluted DNA, 1.5 µL of each primer (10 µM), and water to a total volume of 20 µL. Thermal cycling parameters were as described in Savić and Tibell (Citation2009) and touchdown PCR as in Matheny (Citation2005) for TEF1. Amplification products were visualized on 1.5% agarose gels stained with gel red, and PCR products were purified using Illustra ExoStar buffer (GE Healthcare, UK) diluted 10×, following the manufacturer’s protocol. Sequencing was carried out by Macrogen (www.macrogen.com).
Alignments and phylogenetic analyses.—
Additional sequences from GenBank () were chosen to reconstruct a multigene alignment including as much taxonomic coverage of Fomitopsidaceae and Laetiporaceae following Han et al. (Citation2016) and Song et al. (Citation2018). Alignments were performed using MAFFT 7 online (https://mafft.cbrc.jp/alignment/server/; Katoh et al. Citation2019) and viewed and manually adjusted, where necessary, using AliView 1.26 (Larsson Citation2014). Ambiguously aligned regions were excluded prior to phylogenetic analyses. We retained only the 5.8S region of ITS for the combined data set, since the neighboring regions (ITS1 and ITS2) were poorly aligned. A conflict among single-locus data sets was considered significant if a well-supported monophyletic group (posterior probability [PP] ≥0.95) was found to be well supported as nonmonophyletic when different loci were used. Further analyses were carried out after concatenation using SequenceMatrix (Vaidya et al. Citation2011).
Table 1. Species, sample numbers, and GenBank accession numbers of sequences used in this study; new sequences in bold
The best-fit model of DNA evolution for the analyses, for both individual codon positions and genes, was obtained using Akaike information criterion (AIC) as implemented in MrModeltest 2.3 (Nylander Citation2004). The GTR+I+G model was employed across sites for 28S, 18S, and the 2nd codon of TEF1. For the 1st codon of TEF1, the model F81+I+G was applied, whereas HKY+G was implemented for both the 3rd codon of TEF1 and 5.8S. Bayesian inference (BI) was conducted using MrBayes 3.2.6, and branch support was estimated by the posterior probability (PP) (Ronquist and Huelsenbeck Citation2003; Ronquist et al. Citation2012). Two independent runs were executed, each with four Markov chains for 10 million generations, sampling trees every 100 generations. A 25% threshold was used as the burn-in. The Markov chain Monte Carlo (MCMC) analysis converged well in advance of the burn-in threshold, and chain mixing was found to be satisfactory as assessed by using Tracer 1.5 (Drummond et al. Citation2012).
Maximum likelihood (ML) estimates were carried out by RAxML 8.2.10 using the GTR+G+I model of site substitution (Stamatakis Citation2014). Branch support was obtained by bootstrapping 1000 replicate data sets (Hillis and Bull Citation1993). Sequence alignments and phylogenetic trees were deposited in TreeBASE (submission ID 26318).
RESULTS
Phylogenetic analyses.—
A total of 12 new sequences from three specimen vouchers were generated (). BLAST results from GenBank (accessed 23 Apr 2019), using BLASTn with “discontiguous megablast” (for cross-species comparison, searching with coding sequences) with sequences in this study, showed the highest sequence similarity to Piptoporus soloniensis (ITS query cover 97% and identity 76%).
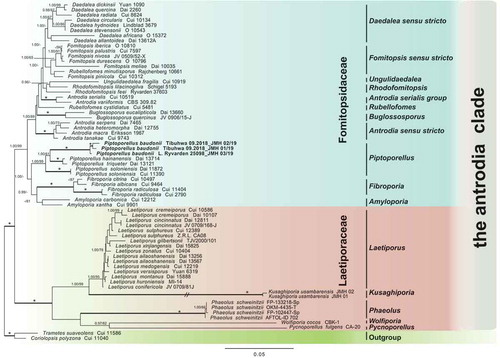
The analyses contained a total of 249 sequences representing 64 species of the “antrodia clade,” including the additional sequences downloaded from GeneBank and two species from the “core polyporoid” clade as outgroups. No significant incongruence among single gene trees was detected; hence, the four matrices were concatenated. The concatenated data matrix contained 2859 unambiguously aligned sites. The phylogeny of the “antrodia clade” and the position of Piptoporellus baudonii were inferred from four data sets: 58 sequences of 5.8S, 66 of 28S sequences, 63 of 18S, and 62 sequences of TEF1. The combined analysis recovered a phylogeny with two distinct clades representing Fomitopsidaceae and Laetiporaceae (). Clade annotations follow Han et al. (Citation2016). Three samples of Laetiporus baudonii clustered (shown as Piptoporellus baudonii, comb. nov., see below) with several samples of Piptoporellus, including the type of the genus P. soloniensis, in the Fomitopsidaceae. This arrangement was strongly supported and necessitated the new taxonomic placement of Laetiporus baudonii in Piptoporellus.
Figure 2. ML tree of Piptoporellus and related genera in the “Antrodia clade” inferred from a combined data set of 5.8S+28S+18S+TEF1. The tree was rooted with two species from the Polyporaceae. Branches are labeled with ML bootstrap values >50% and BPP values >0.95. Branches in bold indicate a support of PP ≥0.95 and boostrap ≥70%. An asterisk above a bold branch indicates 100% bootstrap and 1.0 PP. The branch with a double-slash is shortened. Clade names follow Han et al. (Citation2016)
TAXONOMY
Piptoporellus baudonii (Pat.) Tibuhwa, Ryvarden & S. Tibell, comb. nov.
MycoBank MB835378
≡ Polyporus baudonii Pat., Bull Soc Mycol Fr 30:337. 1914 (basionym).
≡ Pseudophaeolus baudonii (Pat.) Ryvarden, Trans Br Mycol Soc 65:285. 1975.
≡ Laetiporus baudonii (Pat.) Ryvarden, Syn Fung (Oslo) 5:215. 1991.
Basidiomata 20–35 cm wide, pileate, stipitate, and seldom effused-reflexed forming shelves clustered in rosettes up to 85(–110) cm wide, total weight of up to 15 kg; upper surface of pileus bright orange-yellow (5A5–6) when young and fresh, rusty brown (7DE7) upon aging, surface soft without crust, with a few faint concentric brown zones. When aging the basidiomata undergo autolysis. Hymenophore poroid, pore surface concolorous with the upper surface of the pileus or slightly paler. Context fleshy, light ochraceous (2A4–5).
Hyphal system dimitic. Generative hyphae thin-walled, 2–3.5 µm wide, hyaline, septate with clamp connections. Binding hyphae relatively thick-walled, 2–3 µm wide, hyaline, nonseptate with arbori-shaped branching. Basidiospores broadly ellipsoid, or rarely ovoid, 6–7(–7.5) × (3–)3.5–4(–4.5) μm, ave. = 6.5 ± 0.34 × 3.75 ± 0.25 μm, smooth, thin-walled, hyaline, inamyloid. Basidia clavate, (7–)12.5–17.5(–23) × 6.5–7 µm, hyaline, thin-walled, with 2–4 sterigmata. Cystidia not found.
Ecology and distribution: On a wide range of hosts (); for additional host trees, see Van der Westhuizen (Citation1973) and Rattan and Pawsey (Citation1981). Basidiomata occurring during the rainy season (Nov–May). Widely distributed in Africa and Madagascar (Kile Citation2000); also recorded from Yemen (Al-Fatimi et al. Citation2005).
Table 2. Host tree range for Piptoporellus baudonii
Specimens examined: TANZANIA. MTWARA PROVINCE: Naliendele cashew farm, on wilting cashew tree in the vicinity of other wilting eucalyptus trees in cleared farm, 17 Apr 2011, Tibuhwa 1067.2011. (UPS, UDSM); Naliendele cashew farm, 25 Apr 2014, Tibuhwa 1096.2014 (UDSM); Naliendele, in Eucalyptus artificial plantations, 30 Apr 2014, Tibuhwa 1098.2014 (UDSM); Kitangari, 10°37′18.6″S, 39°20′19.5″E, 360 m, growing on the roots of wilting cashew tree, 9 Jan 2018, S. Amandus, Tibuhwa 09.2018 (UDSM; DNA isolations: JMH 01/19, JMH 02/19). GenBank: ITS = MT447066, MT447067; 28S = MT447069, MT447070; 18S = MT447063, MT447064; TEF1 = MT452549, MT452550. ZIMBABWE. CENTRAL PROVINCE: Mazowe Botanic Reserve, ca. 20 km north of Harare, on the ground, 16 Jan 1988, L. Ryvarden 25098 (O; DNA isolation JMH 03/19). GenBank: ITS = MT447068; 28S = MT447071; 18S = MT447065; TEF1 = MT452551. SENEGAL. CAP-VERD: 13 Oct 1984, J. Moen (O). UGANDA: Rwenzori Mountains, 2600 m, on liana, 10 Nov 2007, C. Decock (O). GHANA. GREATER ACCRA: locality: Cassava Farm, on tuber of Cassava, Jan 2011, Mary M. Apetergbor (O).
Remarks: The edibility is unknown, although it was once found eaten by a wild tortoise ().
DISCUSSION
Laetiporus baudonii has had a controversial taxonomic history regarding its generic placement and species recognition as reported previously by Van der Westhuizen (Citation1973). Here, we infer the phylogenetic position of the species using molecular data from four gene regions. In our analyses, L. baudonii from Zimbabwe clustered with two isolates from Tanzania to form a highly supported and distinct species-level lineage within Piptoporellus of the antrodia clade (). Phylogenetic evidence thus justifies the incorporation of Laetiporus baudonii in Piptoperellus along with other sampled members of the genus—P. soloniensis, P. hainanensis, and P. triqueter. Morphologically, P. baudonii is similar to other Piptoporellus in having annual, pileate, or stipitate basidiomata, similar pileus coloration, a dimitic hyphal system, thin-walled basidiospores, and presence of clamp connections. However, P. baudonii differs from the other Piptoporellus species by spore shape (Han et al. Citation2016).
For more than 10 years, farmers in Mtwara, Tanzania, have noticed fungal infections in plantations of cashew. Farmers also observed that when the fungus was found beneath a tree, the leaves of the tree were chlorotic and completely wilted. Defoliation and wilting of the attacked trees continued until the whole tree succumbed, with the wilt spreading to adjacent trees. However, P. baudonii has low host specificity. During our study, we observed its occurrence on a wide range of trees, including Eucalyptus, cashew tree, cassava trees, Tectona trees, and indigenous trees, in southern Tanzania (see ). Researchers at the Naliendele Agricultural Research Institute, Mtwara, have confirmed these observations. A clear indication of local fungal infection was seen in plantations of cultivated Tectona grandis and Eucalyptus trees, where distinct patches of dry and dying trees were noted. Pathogenicity studies of P. baudonii and possible mitigation measures are ongoing.
Piptoporellus baudonii has a deep impact on agroforestry and ecology of the affected areas. Cashew is the main cash crop for more than 380 000 households in southeastern Tanzania, whereas cassava is their major staple food. Infection by P. baudonii, which attacks both their main cash crop and main staple food apart from other cultivated and wild forest trees, is of great concern in this region and a threat to agroforestry. There is concern that a collapse in this system would entail complex and multifarious socioeconomic problems. Mitigation procedures are essential to inhibit spread of the disease caused by P. baudonii.
Supplemental Material
Download (190.3 KB)ACKNOWLEDGMENTS
Naliendele Agriculture Research institute is acknowledged for assisting in field surveys. We thank the editors, Dr. Patrick B. Matheny and Dr. Daniel Linder, and comments from two anonymous reviewers.
Supplemental Material
Supplemental data for this article can be accessed on the publisher’s website.
Additional information
Funding
LITERATURE CITED
- Al-Fatimi M, Wurster M, Kreisel H, Lindequist U. 2005. Antimicrobial, cytotoxic and antioxidant activity of selected basidiomycetes from Yemen. Pharmazie 60:776–780.
- Binder M, Hibbett DS, Larsson KH, Larsson E, Langer E, Langer G. 2005. The phylogenetic distribution of resupinate forms across the major clades of mushroom‐forming fungi (Homobasidiomycetes). Systematics and Biodiversity 3:113–157.
- Browne FG. 1968. Pests and diseases of forest plantation trees. An annotated list of the principal species occur ring in the British Commonwealth. Oxford, UK: Clarendon Press. 923 p.
- Chen Y-Y, Cui B-K. 2016. Phylogenetic analysis and taxonomy of the Antrodia heteromorpha complex in China. Mycoscience 57:1–10.
- Chen Y-Y, Li H-J, Cui B-K. 2015. Molecular phylogeny and taxonomy of Fibroporia (Basidiomycota) in China. Phytotaxa 203:47–54.
- Cui B, Dai Y. 2013. Molecular phylogeny and morphology reveal a new species of Amyloporia (Basidiomycota) from China. Antonie van Leeuwenhoek 104:817–827.
- Donk MA. 1960. The generic names proposed for Polyporaceae. Persoonia 1:173–302.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973.
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Molecular Ecology 2:113–118.
- Han ML, Chen YY, Shen LL, Song J, Vlasák J, Dai YC, Cui BK. 2016. Taxonomy and phylogeny of the brown-rot fungi: Fomitopsis and its related genera. Fungal Diversity 80:343–373.
- Han ML, Cui BK. 2015. Morphological characters and molecular data reveal a new species of Fomitopsis (Polyporales) from southern China. Mycoscience 56:168–176.
- Han M, Song J, Cui B. 2014. Morphology and molecular phylogeny for two new species of Fomitopsis (Basidiomycota) from South China. Mycological Progress 13:905–914.
- Han ML, Vlasák J, Cui BK. 2015. Daedalea americana sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Phytotaxa 204:277–286.
- Heim R. 1931. Le Phaeolus manihotis sp. nov., parasite du manioc a Madagascar, et considérations sur le genre Phaeolus Pat. Annales de Cryptogamie Exotique 4:175–189.
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42:182–192, http://mafft.cbrc.jp/alignment/server/
- Hussein JM, Tibuhwa DD, Tibell S. 2018. Phylogenetic position and taxonomy of Kusaghiporia usambarensis gen. et sp. nov. (Polyporales). Mycology 9:136–144.
- Justo A, Miettinen O, Floudas D, Ortiz-Santana B, Sjökvist E, Lindner D, Nakasone K, Niemelä T, Larsson KH, Ryvarden L, Hibbett DS. 2017. A revised family-level classification of the Polyporales (Basidiomycota). Fungal biology 121:798–824.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20:1160–1166.
- Kirk PM, Cannon PF, Minter DW, Stalpers JA. 2008. Dictionary of the Fungi. 10th ed. Wallingford, UK: CABI. 784 p.
- Kile GA. 2000. Woody Root Rots of Eucalypts. In: Keane PJ, Kile GA, Podger FD, Brown BN, eds. Diseases and pathogens of eucalypts. Melbourne, Australia: CSIRO Publishing. p. 293–303.
- Kim KM, Lee JS, Jung HS. 2007. Fomitopsis incarnatus sp. nov. based on generic evaluation of Fomitopsis and Rhodofomes. Mycologia 99:833–841.
- Kornerup A, Wanscher JH. 1962. Farver i Farver. Copenhagen, Denmark: Politikens Forlag.
- Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278.
- Li H-J, Cui B-K. 2013. Two new Daedalea species (Polyporales, Basidiomycota) from South China. Mycoscience 54:62–68.
- Matheny PB. 2005. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Molecular Phylogenetics and Evolution 35:1–20.
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, Langer E, Langer G, McLaughlin DJ, Wilson AW, Frøslev T, Ge Z-W, Kerrigan RW, Slot JC, Yang Z-L, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vauras J, Hibbett DS. 2007. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 43:430–451.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- Ofosu-Asiedu A. 1975. A new disease of eucalypts in Ghana. Transactions of the British Mycological Society 65:285–289.
- Ortiz-Santana B, Lindner DL, Miettinen O, Justo A, Hibbett DS. 2013. A phylogenetic overview of the antrodia clade (Basidiomycota, Polyporales). Mycologia 105:1391–1411.
- Patouillard NT. 1914. Quelques champignons du Congo. Bulletin de la Société mycologique de France 30:336–346.
- Rattan PS, Pawsey RG. 1981. Death of tea in Malawi caused by Pseudophaeolus baudonii. International Journal of Pest Management 27:225–229.
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574.
- Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MRBAYES 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Systematic Biology 61:539–542.
- Ryvarden L. 1991. Genera of polypores, nomenclature and taxonomy. Synopsis Fungorum (Oslo) 5:1–373.
- Savić S, Tibell L. 2009. Taxonomy and species delimitation in Sporodictyon (Verrucariaceae) in Northern Europe and the adjacent Arctic—reconciling molecular and morphological data. Taxon 58:585–605.
- Sijaona MER. 2003. Pathology report. In: Cashew Research Coordinating Committee Meeting Report, 2003/2004. Naliendele, Mtwara: Agricultural Research Institute. 112 p.
- Sijaona MER. 2007. Macro-fungi in association with cashew wilt disease in Tanzania. A poster presented at the 18th Congress of the Association for the Taxonomic Study of the Flora of Tropical Africa [AETFAT], 26 Feb to 2 Mar 2007, Yaounde, Cameroon.
- Sijaona MER, Shija B. 2005. Verification of Bayleton 25WP for the control of powdery mildew (Oidium anacardii F. Noack) on cashew in Tanzania. Registration report submitted to Registar of Pesticides. Arusha, Tanzania: Tropical Pesticides Research Institute (TPRI).
- Song J, Sun YF, Ji X, Dai YC, Cui BK. 2018. Phylogeny and taxonomy of Laetiporus (Basidiomycota, Polyporales) with descriptions of two new species from western China. MycoKeys 37:57–71.
- Song J, Cui B. 2017. Phylogeny, divergence time and historical biogeography of Laetiporus (Basidiomycota, Polyporales). BMC Evolutionary Biology 17:1–12.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313.
- Tibuhwa DD, Kivaisi AK, Magingo FFS. 2010. Utility of the macro-micromorphological characteristics used in classifying the species of Termitomyces. Tanzania Journal of Science 36:3–46.
- Thiers B. [continuously updated]. Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. [cited 2020 May 20]. Available from: http://sweetgum.nybg.org/science/ih/
- Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27:171–180.
- Van der Westhuizen GCA. 1973. Polyporus baudoni Pat. on Eucalyptus spp. in South Africa. Bothalia 11:143–151.
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172:4238–4246.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, eds. PCR protocols: a guide to methods and applications. New York: Academic Press. p. 315–322.
- Zhao CL, Cui BK, Song, J, Dai YC. 2015. Fragiliporiaceae, a new family of Polyporales (Basidiomycota). Fungal Diversity 70:115–126.
