Abstract
The genetic diversity of 27 wild alfalfa (Medicago sativa L.) populations from seven different countries (Iran, US, Australia, France, Italy, Turkey and Kazakhstan) was studied using total protein profiles and phenotypic traits. High genetic variation was observed for both total protein profiles and phenotypic traits. AMOVA using total protein profiles revealed no variation between local and exotic populations; however, variance between populations and within populations accounted for 33% and 67% of the total variation, respectively. UPGMA was conducted using total protein profiles. All local and exotic wild populations were more or less scattered indicating shared protein profiles that might be due to common parentage or gene flow. The protein banding data were investigated in relation to phenotypic traits and indicated no influence of polymorphic bands on quantitative traits. Latitude, longitude and altitude of origin were not correlated with either genetic or phenotypic distances among Iranian populations.
Introduction
Alfalfa (Medicago sativa L.) is an excellent source of protein, vitamins and minerals, and is the most important forage crop species in Iran. It is also one of the most important forage crops in the world, with about 32 million hectares planted globally. The plant is believed to have originated in the Caucasus region – north-eastern Turkey, Turkmenistan and north-western Iran (Michaud et al. Citation1988). Alfalfa is an autotetraploid (2n =4x=32), cross-pollinated (allogamous), seed-propagated species. These factors contribute to genetic complexity of alfalfa at both individual and population levels (Labombarda et al. Citation2000). The most frequent breeding methods applied to alfalfa involve different forms of mass selection, recurrent phenotypic selection and development of synthetic populations. Information about germplasm diversity and relationships among elite breeding materials is of fundamental importance in plant breeding (Hallauer & Miranda Citation1988). This is especially true for species such as alfalfa which suffers severe inbreeding depression (Bingham et al. Citation1994). Genetic diversity of initial selection materials is essential for successful breeding and creation of new cultivars. For estimates of genetic diversity, criteria such as morphological, agronomic and physiological characteristics, pedigree records, molecular markers or a combination of these are used. Alfalfa is distributed worldwide and grown in highly contrasting environments. This wide geographical adaptation enhances genetic variation and provides the opportunity to use diverse gene pools in breeding programmes (Tucak et al. Citation2008). Studies conducted on alfalfa in different ecological conditions in Iran revealed that there was considerable variation in herbage yield, seed yield and crude protein content (Jafari & Goodarzi Citation2007).
The introduction of biochemical and molecular techniques has allowed a more accurate evaluation of genetic relationships of alfalfa: allozyme gene markers (Fayd-Lameche et al. Citation1996; Jenczewski et al. Citation1999), total protein profiles (Krochko & Bewley Citation2000; Chandra & Panday Citation2011; Radović et al. Citation2012), restriction fragment length polymorphism (RFLP) (Maureira et al. Citation2004), random amplified polymorphic DNA (RAPD) (Tucak et al. Citation2008), simple sequence repeat (SSR) (Falahati-Anbaran et al. Citation2007), sequence related amplified polymorphisms (SRAP) (Vandemark et al. Citation2005) and amplified fragment length polymorphism (AFLP) (Keivani et al. Citation2010). However, the protein profiling of germplasm and the use of genetic markers have been widely and effectively used to determine the taxonomic and evolutionary aspects of several crops (Khan Citation1990; Murphy et al. Citation1990; Das & Mukharjee Citation1995; Ghafoor et al. Citation2002; Emre et al. Citation2007; Radović et al. Citation2012). Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) is the most economical, simple and extensively used biochemical technique for analysis of genetic structure of germplasm. Leaf total proteins have been used as genetic markers in breeding programmes (Hirano Citation1982; Vries Citation1996; Kamel & Hassan Citation2001; Reddy & Munirajappa Citation2005; Mohamed et al. Citation2006; Chandra & Panday Citation2011), as well as in basic studies on population genetics (Torkpo et al. Citation2006) and reproductive biology (Cardeña et al. Citation1998). To our knowledge, no studies have yet been made in Iran on the diversity of alfalfa germplasm based on total protein electrophoresis, and its association with phenotypic traits.
Erosion of diversity in most cultivated species emphasises the need to collect and investigate new germplasms as genetic resources for future breeding programmes. Alfalfa is distributed worldwide and grown in highly contrasting environments. This extensive geographical adaptation promotes genetic variation and gives us the opportunity to use diverse gene pools. The aims of this research were to study genetic diversity among alfalfa populations with different geographical origin (Iran, US, Australia, France, Italy, Turkey and Kazakhstan) using total protein profiles and morphological traits, to determine genetic variation among and within populations using total protein profiles, and to compare results based on molecular markers and morphological traits.
Materials and methods
Seed material and experiment layout
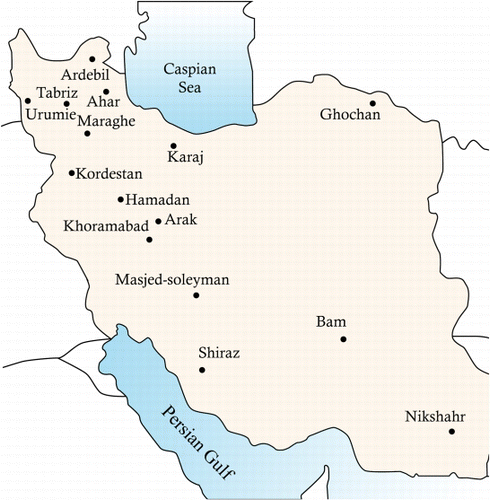
Seed material of 27 populations of alfalfa from Iran (21 populations, ), US, Australia, France, Italy, Turkey and Kazakhstan, provided from the Iranian Natural Resources Gene Bank at the Research Institute of Forests and Rangelands (RIFR), was evaluated in the present study. All the materials were non-dormant.
The research was conducted on the experimental field at the RIFR. A total of 30 seedlings of each population were grown in jiffy pots for 40 days before being transplanted into the field in April 2007. The field trial was arranged in a randomized complete block with three replications. Each plot included 36 spaced plants (0.40×0.40 m). Phosphorus (P) fertilizer was applied at 100 kg/ha at sowing. The field was irrigated once a week during summer. No measurements were taken in the establishment year. Three yield cuts per year were performed during the growing season in 2008 and 2009.
During the two-year investigation, seven phenotypic traits were observed in this research. The following measurements were taken on all individual plants of each population in all six cuts: dry matter yield, day to flowering, regrowth rate, crown diameter, tiller number, plant height and leaf/stem weight ratio. Prior to cuts, plant height (cm) was measured from the ground to the top of the inflorescence. Regrowth rate was assessed within 2 weeks after the previous cut by individual measurements of plant height. Tiller numbers of individual plants were recorded directly, following cuts. For determination of dry matter yield (kg/ha), green yield was determined and samples were dried in ovens at 70 °C to a constant weight (Martin et al. Citation1990). Dried samples were hand-separated into leaf and stem and weighed to determine leaf/stem weight ratio. Crown diameter was measured individually (cm) for each plant. The dry matter yield was the total amount of three cuts per year, whereas the other agronomical traits were averaged based on three cuts per year. Data were also subjected to a combined analysis of variance across years using a split-plot-in-time design with years as sub-plots (Steel & Torrie Citation1980). The data presented in the current study are average values over 2 years.
All populations were tested for germination characteristics. The normal ISTA (Citation1993) laboratory germination test procedure was used with three replications. Seeds (150) of each population were sterilized with 70% ethyl alcohol for 5 min, and then washed with distilled water. Three replicates (50 seeds per replicate) of sterilized seed were placed in Petri dishes on double Whatman papers (TP). For protection against moulds, the water used to moisten the seed samples and substrata contained 0.002% Benomyl fungicide. The samples were transferred into a germinator at (20±4 °C) with 1000 lux light for 15 days. The percentage and speed of germination were recorded at 3, 6, 9, 12 and 15 days. The length of roots and shoots (mm) of 10 randomly selected seedlings (15-day-old) from each replicate were measured. After measuring shoot and root lengths, the caryopses were cut from the seedlings and fresh seedling weights of each replicate were recorded. The seedlings were then placed in an oven at 80 °C for 24 hours, after which the dry weight of each replicate was recorded as a percentage of the fresh weight. Seed vigour index was calculated by multiplying germination (%) and seedling length (Abdul-Baki & Anderson Citation1973).
Total protein analysis
For study of the extent of genetic variation based on SDS-PAGE markers, a total of 270 entries were selected from 27 local and exotic populations (10 plants for each population). Preliminary experiments (data not shown) indicated that a larger sample (20 plants for each population) did not substantially modify the results with respect to the structure of polymorphism. Total proteins were extracted from 14-day-old seedlings using protein extraction 0.05M Tris-HCl, pH=8, 0.2% SDS, 5M urea, 1% B-mercaptoethanol. Electrophoresis was carried out in the discontinuous SDS-PAGE system of Laemmli (Citation1970) using 12% (w/v) separating gel and 5% (w/v) stacking gel. The molecular weights of the dissociated proteins were estimated by using molecular weight standard proteins ‘MW-SDS-70 Kit’. Gels were shaken gently until the background of the gel became clear and polypeptide bands were clearly visible.
Statistical analysis
Analysis of variance was computed on the collected data for each trait for morphological and seed germination traits. The descriptive statistics and phenotypic correlation coefficients between traits were estimated using the SAS9.1 software (SAS Institute Inc. 2003). Thirteen classification variables had significant (P≤0.01) variation among the population and were subsequently used for multivariate analysis. The Euclidean distances of populations were computed on phenotypic traits and then they were used for UPGMA (unweighted pair group method with arithmetic means) cluster analysis using computer package NTSYS-pc (numerical taxonomy and multivariate analysis system) (Rohlf Citation2004).
For protein profile data, to avoid taxonomic weighing, the intensity of bands was not taken into consideration, only the presence of bands was taken as indicative. The scores were 1 for the presence and 0 for the absence of a band. Then, the indices of genetic diversity, such as the percentage of polymorphism and expected heterozygosity (He), were calculated using POPGENE 32 software (Yeh et al. Citation1999). At the same time, the genetic structure within and among populations was detected using the software AMOVA-PREP1.01 (Miller Citation1997) and WINAMOVA (Excoffier Citation1995) in order to partition the genetic variation between local and exotic groups, among populations and within populations. The significance of each variance component was tested with permutation tests (Excoffier et al. Citation1992). Genetic distances were estimated according to Nei (Citation1978) and the resulting distance matrix was subjected to principal component analysis (PCA) and algorithm of UPGMA using NTSYS-pc 2.01 (Rohlf Citation2004). The Mantel test (Gower Citation1966) was used to assess the correlation between the calculated distance matrices (using phenotypic and total protein profiles data), and a permutation procedure was used to assess the significance of this correlation.
Results
Phenotypic data
Basic descriptive statistics for the morphological traits of the local and exotic populations are shown in . Pearson correlation showed a positive relationship between dry matter yield with day to flowering, regrowth rate, tiller number and plant height. Germination percentage positively correlated with day to flowering, speed of germination, seedling length, seed vigour index and seedling root/shoot length ratio, while a negative value was obtained between germination percentage and leaf/stem weight ratio and seedling dry/fresh weight ratio ().
Table 1 . Descriptive statistics for the phenotypic traits in the 27 local and exotic populations of M. sativa.
Table 2 . Pearson correlation analysis for the relationships between phenotypic and genetic parameters of the 27 local and exotic populations of M. sativa.
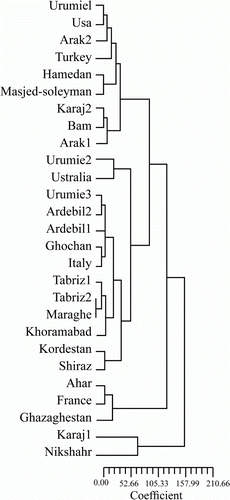
Genetic distance among the 27 alfalfa populations was also estimated using data on 13 phenotypic traits using Euclidean distances among populations and ranged from 1.00 (between Karaj and Shiraz, both from Iran) to 9.96 (between Uromieh2 from Iran and Turkey) with an average value of 3.87 (). The Euclidean distances matrix was subjected to agglomerative hierarchical clustering utilizing the UPGMA method to construct a dendrogram (). Twenty-seven populations of alfalfa were classified into three groups. Cluster I consisted of 22 populations, cluster II included populations Ahar from Iran, France and Kazakhstan, and cluster III consisted of populations Karaj1 and Nikshahr, both from Iran (). Therefore, there was no relationship between phenotypic traits and the origin of these alfalfa populations.
Table 3 . Pair-wise values for squared Euclidean distances (below diagonal) and Nei's genetic distances (above diagonal) of the 27 local and exotic populations of M. sativa.
Total protein profiles
On the basis of the relative mobility of total proteins on the gel, 44 polypeptide bands of different sizes ranging from 6.606 to 269.153 kDa, from 27 populations of alfalfa, were identified. The percentages of polymorphic bands over the total bands detected ranged from 6.82% (Karaj1 and Urumie2, both from Iran) to 40.91% (Maraghe and Ahar, both from Iran) with an average of 25.51% (). Assuming Hardy–Weinberg equilibrium, the value of Nei's genetic diversity (He) ranged from 0.030 (Karaj1 from Iran) to 0.172 (Ahar from Iran) (). High polymorphism was found within populations and the probability that two randomly sampled polypeptides in a given populations are different was 9.7% (He=0.097).
Table 4 . Genetic diversity parameters of the 27 local and exotic populations of M. sativa.
AMOVA using total protein profiles revealed no variation among local and exotic groups, however variance among populations and within populations accounted for 33% and 67% of the total variation, respectively (). This difference was statistically significant (P<0.01) based on the permutation test.
Table 5 . Analysis of molecular variance (AMOVA) of the 27 local and exotic populations of M. sativa.
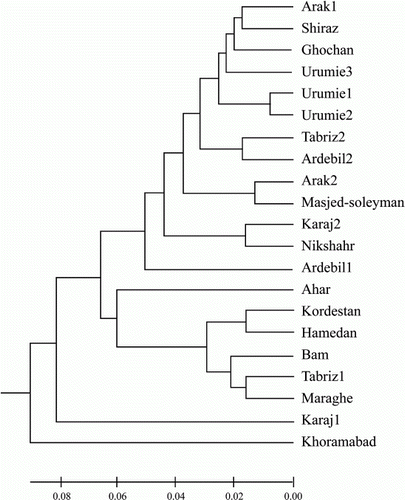
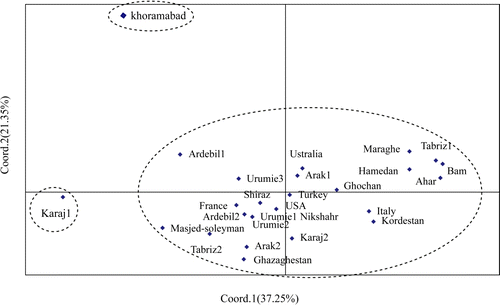
The pairwise values for Nei's genetic distances between the analysed populations ranged from 0.005 (between Arak2 from Iran and Kazakhstan) to 0.241 (Khoramabad and Ahar, both from Iran), with an average of 0.069 (). To elucidate the genetic relationships between alfalfa populations, an emphasized UPGMA dendrogram was produced using Nei's genetic distances (). The 27 populations were grouped into three clusters, cluster I having 25 populations, cluster II consisted of population Karaj1, and cluster III also having only one population (Khoramabad). The total protein data were also used for conducting PCA to study further the genetic diversity among the 27 alfalfa populations (). The first three components of PCA accounted for 71.79% of the total variation. The 27 populations were separated into three groups, the smaller groups each comprising one population and the larger group comprising 25 populations. This clustering pattern of genotypes obtained on the basis of PCA was similar to the clustering of populations in the dendrogram obtained through UPGMA analysis (compare and ). Geographic origin was not related to genotypic similarity among populations. However, there were instances of close genetic relatedness between accessions from neighbouring locations in a geographic compartment. In other words, this clustering pattern, made on the basis of SDS-PAGE, grouped the accessions differently and gave no clear indication of phenotypic performance or origin/source.
Correlation between genetic and phenotypic traits and ecological factors
Correlation coefficients among pairwise genetic and phenotypic distance matrices were calculated using Mantel's test. Regression and correlation analysis between genetic and phenotypic distances showed no significant correlation (P>0.05).
The correlation analysis () indicated that He of different Iranian populations had no significant correlation with elevation and climate factors. However, some phenotypic traits had significant correlation with elevation and climate factors. The day to flowering and germination percentage had positive correlation with elevation. These two indices also had a significant correlation (negative) with average annual maximum and minimum temperatures. Five out of six germination indices, the speed of germination, germination percentage, seedling height, seed vigour index and seedling root/shoot length ratio, showed positive correlation with average annual precipitation. The above correlations implied that the genetic diversity of alfalfa was not the result from the joint effects of one or several ecological factors, i.e. the ecological factors have not played an important role in influencing the protein profiles polymorphism of alfalfa.
Table 6 . Pearson correlation analysis for the relationships between ecological factors and phenotypic and genetic parameters of the Iranian populations of M. sativa.
The Pearson correlation data showed significant correlation (P>0.01) between He and plant height.
Discussion
In the present investigation, high genetic variation was observed for total protein profiles and phenotypic traits. Parallel to our findings, significant variations were observed with respect to morphological, phenological, biological and molecular properties between populations in previous studies (Maureira et al. Citation2004; Jafari & Goodarzi Citation2007; Tucak et al. Citation2009; Benabderrahim et al. Citation2009, Citation2011; Xavier et al. Citation2011). The reason for this variation detected within populations may be related to genetic structure, which is probably due to heterozygosis of cross-pollination of alfalfa (Prosperi et al. Citation2006). The cross-pollination mechanism, sexual reproduction, high seed ratio and incompatibility to produce offspring of the Medicago species could have resulted in the accumulation of abundant genetic variation during its long evolutionary history (Xavier et al. Citation2011). This indicates that improvement through simple selection for these traits is possible. However, broadening the genetic base from diverse sources is recommended to include most of the genetic determinants of these traits (Laghetti et al. Citation1998; Ghafoor et al. Citation2002).
Protein profile variation revealed that alfalfa populations held more genetic variation within rather than among populations (67% and 33%, respectively). According to Hamrick & Godt (Citation1996), reproductive biology is the most important factor in determining the genetic structure of plant populations. They showed that out-crossing plant species tend to exhibit between 10% and 20% genetic variation among populations, while self-pollination species exhibit on average 50% variation among populations. Therefore, the 33% genetic variation among studied populations can be partially explained by inbreeding. Although studies on the biology of flowering and pollination in alfalfa indicate it as an out-crosser (Barcaccia et al. Citation1996), alfalfa shows partial self-compatibility. Lv et al. (Citation2009) studied the characteristics of self-compatibility and pollination approaches in eight alfalfa cultivars from different sources and found the self-compatibility of alfalfa differed with cultivar. Wang et al. (Citation2009) also showed that there was significant variation in the self-compatibility rate of alfalfa. Zhang et al. (Citation2006) reported that flowering behaviour and tripping phenomenon in M. lupulina depended on the weather conditions, so that temperature and illumination are important factors affecting tripping. Mable & Adam (Citation2007) reported in Arabidopsis lyrata, which is usually considered an obligatory out-crossing species with a strong self-incompatibility system, that a shift in mating systems towards inbreeding has been found in some populations. A common prediction, particularly in the conservation literature, is that a shift from out-crossing to inbreeding could be an adaptive strategy to deal with habitat fragmentation and/or reduced population size resulting from human activities or geological change (Hedrick & Kalinowski Citation2000; Picó et al. Citation2003; Zoro et al. Citation2003). It is thus difficult to draw conclusions about whether recent habitat fragmentation has favoured or promoted the loss of self-incompatibility in these populations, but current population density appears to be a major factor, because wild populations of M. sativa L. from Iran and also many other countries occur along roadsides and fields.
AMOVA showed significant differences among populations but not between the local and exotic populations. It revealed that geographic origin was not related to genotypic similarity among populations. However, in controversial reports, Mengoni et al. (Citation2000) and Yan et al. (Citation2009) detected the significantly positive correlation between genetic and geographical distances in M. sativa and M. lupulina populations with different geographical origins, respectively. In general they found a tendency for closely situated populations to be genetically more similar. Results of this study showed that genetic diversity and geographic distribution in wild populations of alfalfa were independent of each other and no definite relationship existed between genetic diversity and geographic diversity, which is in accordance with the results of the study on alfalfa using allozymes (Fayd-Lameche et al. Citation1996; Jenczewski et al. Citation1999), total protein profiles (Krochko & Bewley Citation2000; Živković et al. Citation2012), AFLP (Keivani et al. 2010), RAPD (Noeparvar et al. Citation2008; Tucak et al. Citation2008), SSRs (Touil et al. Citation2008); and also other species including Mentha spp. (Badr et al. Citation2003), Lathyrus sativus L. (Sammour et al. Citation2007), Lens spp. (Sultana et al. Citation2006) and Medicago truncatula (Akritidis et al. Citation2009). In spite of other studies with alfalfa using SSRs (Falahati-Anbaran et al. Citation2007; Petolescu et al. Citation2010), or RAPD (Wang et al. Citation2011), that suggest ecological conditions cause the observed variations, the results of this current work imply that the genetic diversity of alfalfa is not the result of joint effects of ecological factors, i.e. the ecological factors have not played an important role in influencing the protein profile polymorphism of alfalfa. This study provides evidence that use of the total protein marker is an informative and suitable approach for evaluation of molecular polymorphism and polygenic relationships in wild populations of alfalfa.
The Mantel's test confirmed the low correlation between total protein profiles and phenotypic trait distances. This is in agreement with studies of other authors who used morphological characters and molecular markers (Crochemore et al. Citation1996; Tucak et al. Citation2008; Ping et al. Citation2009). The result suggests that the two marker systems give different estimates of genetic relations among populations. It may be noted that a small number or common set of genes/quantitative trait loci (QTL) may be controlling several correlated phenotypic traits evaluated in the present study. Therefore, the genome coverage represented by phenotypic traits is likely to be poor, and this may possibly be the reason for poor correlation between the genetic distance values based on phenotypic traits and genetic similarity values based on molecular markers, which represent relatively better genome coverage. This problem, however, may be overcome by using markers developed from expressed sequences that may be directly responsible for the traits related to fitness and adaptation of genotypes. For example, in a recent study on barley, in comparison with SSR markers, the RFLP analysis of stress responsive genes (SRG-RFLP) produced a better separation of barley cultivars according to eco-morphological characteristics (Maestri et al. Citation2002).
Conclusion
From the present study it can be concluded that there was a close genetic relationship between local and exotic alfalfa populations and that great variation exists among populations within local or exotic groups. Genetic diversity and relationships of wild alfalfa can be used in the development of germplasm collection, breeding and conservation. The results clearly indicate that comprehensive germplasm collection in major geographic regions is required to broaden the genetic base and sample the full extent of the available variation. Breeding strategies need to exploit the existing variation within the wild alfalfa germplasm. Also, the study confirmed that genetic and morphological diversity work in different ways to determine the relationships among populations. To effectively exploit germplasms, we should utilize both methods in breeding work. Further studies are required to reveal whether there are other factors that cause genetic variation in alfalfa.
Acknowledgements
This work was supported by the Research Institute of Forests and Rangelands (RIFR), Iran. It was project no. 12-09-09-7901-87001.
References
- Abdul-Baki , AA and Anderson , JD . 1973 . Vigor determination in soybean seed by multiple criteria . Crop Science , 13 : 630 – 633 . doi: 10.2135/cropsci1973.0011183X001300060013x
- Akritidis , P , Mylona , PV , Tsaftaris , AS and Polidoros , AN . 2009 . Genetic diversity assessment in Greek Medicago truncatula genotypes using microsatellite markers . Biologia Plantarum , 53 : 343 – 346 . doi: 10.1007/s10535-009-0063-6
- Badr A , Mustafa A-Z MA , El-galaly MA , Mobarak AA , Hassan MG 2003 . Genetic diversity among menthe populations in Egypt as reflected by morphological and protein electrophoretic variations . Proceedings of the 1st Egyptian and Syrian Conference for Agriculture and Food , El Minia , Egypt , 8 – 11 September 2003 . 269 – 286 .
- Barcaccia , G , Mazzucato , A , Falcinelli , M and Veronesi , F . 1996 . Callose localization in cell walls during meiotic and apomeiotic megasporogenesis in diploid alfalfa . Caryologia , 49 : 45 – 56 .
- Benabderrahim , MA , Haddad , M , Hamza , H and Ferchichi , A . 2011 . Germination and emergence variability of alfalfa (Medicago sativa L.) landraces collected in Southern Tunisia oases . Spanish Journal of Agricultural Research , 9 : 135 – 143 .
- Benabderrahim , MA , Haddad , M and Ferchichi , A . 2009 . Diversity of lucerne (Medicago sativa L.) populations in south Tunisia . Pakistan Journal of Botany , 41 : 2851 – 2861 .
- Bingham , ET , Groose , RW , Woodfield , DR and Kidwell , KK . 1994 . Complementary gene interactions in alfalfa are greater in autotetraploids than diploids . Crop Science , 34 : 823 – 829 . doi: 10.2135/cropsci1994.0011183X003400040001x
- Cardeña , R , Oropeza , C and Zizumbo , D . 1998 . Leaf proteins as markers useful in the genetic improvement of coconut palms . Euphytica , 102 : 81 – 86 . doi: 10.1023/A:1018392908569
- Chandra , A and Panday , KC . 2011 . Assessment of genetic variation in lucerne (Medicago sativa L.) using protease inhibitor activities and RAPD markers . Journal of Environmental Biology , 32 : 559 – 565 .
- Crochemore , ML , Huyghe , C , Kerlan , MC , Durand , F and Julier , B . 1996 . Partitioning and distribution of RAPD variation in the Medicago sativa complex . Agronomie , 16 : 421 – 432 . doi: 10.1051/agro:19960702
- Das , S and Mukharjee , KK . 1995 . Comparative study on seed proteins of Ipomoea . Seed Science Technology , 23 : 501 – 509 .
- Emre , I , Turgut-Balik , D , Sahin , A and Kursat , M . 2007 . Total electrophoretic band patterns of some Onobrychis species growing in turkey . American-Eurasian Journal of Agriculture and Environment Science , 2 : 123 – 126 .
- Excoffier L 1995 . AMOVA 1.55 (Analysis of Molecular Variance) . University of Geneva , Switzerland , Genetics and Biometry Laboratory .
- Excoffier , L , Smouse , P and Quattro , J . 1992 . Analysis of molecular variances among DNA restriction data . Genetics , 131 : 479 – 491 .
- Falahati-Anbaran , M , Habashi , AA , Esfahany , MS , Mohammadi , A and Ghareyazie , B . 2007 . Population genetic structure based on SSR markers in alfalfa (Medicago sativa L.) from various regions contiguous to the centres of origin of the species . Journal of Genetics , 86 : 59 – 63 . doi: 10.1007/s12041-007-0008-9
- Fayd-Lameche , FZ , Bellatar , G , Bouabdallah , S and Yahia , N . 1996 . Between and within species variation in annual Medicago species . Cahiers Options Méditerranéennes , 18 : 161 – 169 .
- Ghafoor , A , Ahmad , Z , Qureshi , AS and Bashir , M . 2002 . Genetic relationship in Vigna mungo (L.) Hepper and V. radiate (L.) R. Wilczek based on morphological traits and SDS PAGE . Euphytica , 123 : 367 – 378 . doi: 10.1023/A:1015092502466
- Gower , JC . 1966 . Some distance properties of latent root and vector methods used in multivariate analysis . Biometrika , 53 : 325 – 338 .
- Hallauer , AR and Miranda , JB . 1988 . Quantitative genetics in maize breeding , 2nd edition , 125 Ames , Iowa : Iowa State University Press .
- Hamrick , JL and Godt , MJW . 1996 . Effects of the history traits on genetic diversity in plants . Philosophical Transactions B , 351 : 1291 – 1298 . doi: 10.1098/rstb.1996.0112
- Hedrick , PW and Kalinowski , ST . 2000 . Inbreeding depression in conservation biology . Annual Review of Ecology and Systematic , 31 : 139 – 162 . doi: 10.1146/annurev.ecolsys.31.1.139
- Hirano , H . 1982 . Varietal differences of leaf protein profiles in mulberry . Phytochemistry , 21 : 1513 – 1518 .
- ISTA 1993 . International rules for seed testing . Seed Science and Technology 21 : 288 .
- Jafari A , Goodarzi A 2007 . Genetic variation for yield and its relationships with quality and agronomic traits in 72 accessions of alfalfa . Iranian Journal of Rangeland and Forest Plant Breeding Genetic Research 14 : 229 – 239 .
- Jenczewski , E , Prosperi , JM and Ronfort , J . 1999 . Evidence for gene flow between wild and cultivated Medicago sativa (Leguminosae) Based on allozyme markers and quantitative traits . American Journal of Botany , 86 : 677 – 687 . doi: 10.2307/2656577
- Kamel , EA and Hassan , LMA . 2001 . The significance of cuticular features, petiole anatomy and SDS-PAGE in the Taxonomy of the Lauraceal . Pakistan Journal of Biological Sciences , 4 : 1094 – 1100 . doi: 10.3923/pjbs.2001.1094.1100
- Keivani , M , Ramezanpour , SS , Soltanloo , H , Choukan , R , Naghavi , M and Ranjbar , M . 2010 . Genetic diversity assessment of alfalfa (Medicago sativa L.) populations using AFLP markers . Australian Journal of Crop Science , 4 : 491 – 497 .
- Khan , MK . 1990 . Production and utility of chickpea (Cicer arietinum L.) in Pakistan . Progressive Farming , 10 : 28 – 33 .
- Krochko , JE and Bewley , JD . 2000 . Seed storage proteins in cultivars and subspecies of alfalfa (Medicago sativa L.) . Seed Science Research , 10 : 423 – 434 .
- Labombarda , P , Pupilli , F and Arcioni , S . 2000 . Optimal population size for RFLP-assisted cultivar identification in alfalfa (Medicago sativa L.) . Agronomie , 20 : 233 – 240 . doi: 10.1051/agro:2000123
- Laemmli , UK . 1970 . Cleavage of structural proteins during the assembly of the head of bacteriophage T4 . Nature , 227 : 680 – 685 . doi: 10.1038/227680a0
- Laghetti , G , Pienaar , BL , Pasdulosi , S and Perrino , P . 1998 . Ecogeographical distribution of Vigna savi in southern Africa and some areas of the Mediterranean basin . Plant Genetic Resource Newsletter , 115 : 6 – 12 .
- Lv , LY , Wei , ZW , Zhao , Y , Geng , XL , Liu , GJ and Wu , ZN . 2009 . A study of self-compatibility and pollinations and separation of offspring traits of alfalfa . Pratacultural Science , 26 : 33 – 36 .
- Mable , BK and Adam , A . 2007 . Patterns of genetic diversity in selfing and outcrossing populations of Arabidopsis lyrata . Molecular Ecology , 16 : 3565 – 3580 . doi: 10.1111/j.1365-294X.2007.03416.x
- Maestri , E , Klueva , N , Perrotta , C , Gulli , M , Nguyen , HT and Marmiroli , N . 2002 . Molecular genetics of heat tolerance and heat shock proteins in cereals . Plant Molecular Biology , 48 : 667 – 681 . doi: 10.1023/A:1014826730024
- Martin , RC , Voldeng , HD and Smith , DL . 1990 . Intercropping soybean for silage in a cool-temperate region: yield, protein and economic effects . Field Crops Research , 23 : 295 – 310 . doi: 10.1016/0378-4290(90)90061-F
- Maureira , IJ , Ortega , F , Campos , H and Osborn , TC . 2004 . Population structure and combining ability of diverse Medicago sativa germplasms . Theoretical and Applied Genetics , 109 : 775 – 782 . doi: 10.1007/s00122-004-1677-x
- Mengoni , A , Gori , A and Bazzicalup , O . 2000 . Use of RAPD and mircosatellite (SSR) variation to assess genetic relationships among populations of tetraploid alfalfa, Medicago sativa . Plant Breeding , 119 : 311 – 317 . doi: 10.1046/j.1439-0523.2000.00501.x
- Michaud R , Lehman WF , Runbaugh MD 1988 . World distribution and historical development . In : Hanson AA , Barnes DK , Hill RR Alfalfa and alfalfa improvement . US, Madison . 25 – 92 .
- Miller MP 1997 . AMOVA-PREP, a program for the preparation of AMOVA input files for use with WINAMOVA . Department of Biological Sciences, Northern Arizona University , Flagstaff , AZ .
- Mohamed , TR , Khalifa , SF and Salah , EL-Dine RM . 2006 . Leaf protein electrophoretic profiles and chromosome numbers of some Araceae . International Journal of Agriculture and Biology , 8 : 231 – 234 .
- Murphy , RW , Sites , JW , Buth , DG and Haufler , CH . 1990 . “ Protein I: Isozyme electrophoresis ” . In Molecular systematics , Edited by: Hillis , DH and Moritz , C . 45 – 126 . Sunderland , MA : Sinauer Assoc .
- Nei , M . 1978 . Estimation of average heterozygosity and genetic distance from a small number of individuals . Genetics , 89 : 583 – 590 .
- Noeparvar , S , Valizadeh , M , Monirifar , H , Haghighi , AR and Darbani , B . 2008 . Genetic diversity among and within alfalfa populations native to Azerbaijan based on RAPD analysis . Journal of Biological Research , 10 : 159 – 169 .
- Petolescu , C , Ciulca , S , Lazar , A , Schitea , M and Badea , EM . 2010 . Intra-population genetic diversity in Romanian alfalfa cultivars as revealed by SSR markers . Romanian Biotech Letters , 15 : 107 – 112 .
- Picó FX , Ouborg NJ , Van Groenendael JM 2003. Fitness traits and dispersal ability in the herb Tragopogon pratensis (Asteraceae): decoupling the role of inbreeding depression and maternal effects . Plant Biology 5 : 522 – 530 .
- Ping , L , Yunwen , W , Xiaolong , S and Jianguo , H . 2009 . Using microsatellite (SSR) and morphological markers to assess the genetic diversity of 12 falcata (Medicago sativa spp. falcata) populations from Eurasia . African Journal of Biotechnology , 8 : 2102 – 2108 .
- Prosperi , JM , Jenczewski , E , Angevain , M and Ronfort , J . 2006 . Morphologic and agronomic diversity of wild genetic resources of Medicago sativa L. collected in Spain . Genetic Resources and Crop Evolution , 53 : 843 – 856 . doi: 10.1007/s10722-004-6476-3
- Radović , BŽ , Sokolović , D , Šiler , B , Banjanac , T and Štrbanović , R . 2012 . Assessment of genetic diversity among alfalfa (Medicago sativa L.) genotypes by morphometry, seed storage proteins and RAPD analysis . Industrial Crops and Products , 40 : 285 – 291 . doi: 10.1016/j.indcrop.2012.03.027
- Reddy PMM , Munirajappa 2005 . Electrophoretic studies in induced mutants of diploid mulberry genotype S13 . Indian Journal of Biotechnology 4 : 422 – 423 .
- Rohlf JF 2004 . NTSYS-pc: Numerical taxonomy and multivariate analysis system , version 2.11 . Exeter, Setauket , NY .
- Sammour , R , Mustafa , A , Badr , S and Taher , W . 2007 . Genetic variations in accessions of Lathyrus sativus L . Acta Botanica Croatica , 66 : 1 – 13 .
- Steel , RGD and Torrie , JH . 1980 . Principles and procedures of statistics. A biometrical approach , 2nd edition , Tokyo , , Japan : McGraw Hill International Book Co .
- Sultana , T , Ghafoor , A and Ashraf , M . 2006 . Geographic patterns of diversity of cultivated lentil germplasm collected from Pakistan, as assessed by seed protein assays. Acta Biologica Cracoviensia, Series Botanica . Poland , 48 : 77 – 84 .
- Torkpo , S , Danquah , E , Offei , S and Blay , E . 2006 . Esterase, total protein and seed storage protein diversity in Okra (Abelmoschus esculentus L.) . West African Journal of Applied Ecology , 9 : 7 – 18 .
- Touil , L , Guesmi , F , Fares , K , Zagrouba , C and Ferchichi , A . 2008 . Genetic diversity of some Mediterranean populations of the cultivated alfalfa (Medicago sativa L.) using SSR markers . Pakistan Journal of Biological Sciences , 11 : 1923 – 1928 . doi: 10.3923/pjbs.2008.1923.1928
- Tucak , M , Popovic , S , Cupic , T , Grljusic , S , Bolaric , S and Kozumplik , V . 2008 . Genetic diversity of alfalfa (Medicago spp.) estimated by molecular markers and morphological characters . Periodicum Biologorum , 110 : 243 – 249 .
- Tucak , M , Popovic , S , Cupic , T , Šimic , G , Gantner , R and Meglic , V . 2009 . Evaluation of alfalfa germplasm collection by multivariate analysis based on phenotypic traits . Romanian Agricultural Research , 26 : 47 – 52 .
- Vandemark GJ , Hughes TJ , Larsen RC 2005 . Genetic similarities between alfalfa cultivars based on sequence related amplified polymorphism (SRAP) DNA markers. Plant and Animal Genomes XIII Conference , San Diego , CA , US , 15–19 January .
- Vries , IM . 1996 . Characterization and identification of Lactuca sativa cultivars and wild relatives with SDS–electrophoresis (Lactuca sect. Lactuca, Compositae) . Genetic Resources and Crop Evolution , 43 : 193 – 202 . doi: 10.1007/BF00123271
- Wang , XL , Yan , XD , Xu , YP , Lin , CQ and Zhao , ZX . 2009 . Study on the factors influenced fecundity of inbred of alfalfa . Journal of Hebei Agricultural Sciences , 5 : 13 – 14 .
- Wang , XJ , Yang , XL , Chen , L , Feng , GH , Zhang , JW and Jin , L . 2011 . Genetic diversity among alfalfa (Medicago sativa L.) cultivars in Northwest China . Acta Agriculturae Scandinavica Section B-Soil and Plant Science , 61 : 60 – 66 .
- Xavier , JR , Kumar , J and Srivastava , RB . 2011 . Characterization of genetic structure of alfalfa (Medicago sp.) from trans-Himalaya using RAPD and ISSR markers . African Journal of Biotechnology , 10 : 8176 – 8187 .
- Yeh FC , Yang RC , Boyle T 1999 . POPGENE version 1.32, Microsoft window base software for population genetic analysis: a quick user's guide . Alberta , , Canada , University of Alberta, Center for International Forestry Research .
- Yan , J , Chu , HJ , Wang , HC , Li , JQ and Sang , T . 2009 . Population genetic structure of two Medicago species shaped by distinct life form, mating system and seed dispersal . Annals of Botany , 103 : 825 – 834 . doi: 10.1093/aob/mcp006
- Zhang , AQ , Zhu , JZ and Tan , DY . 2006 . A study on the effect of environment factors on flowering behavior and tripping of Xinmu No.1 Medicago lupulina . Acta Prataculturae Sinica , 4 : 43 – 50 .
- Živković , B , Radović , D , Sokolović , D , Šiler , B , Banjanac , T and Štrbanović , R . 2012 . Assessment of genetic diversity among alfalfa (Medicago sativa L.) genotypes by morphometry, seed storage proteins and RAPD analysis . Industrial Crops and Products , 40 : 285 – 291 . doi: 10.1016/j.indcrop.2012.03.027
- Zoro , BI , Maquet , A and Baudoin , JP . 2003 . Population genetic structure of wild Phaseolus lunatus (Fabaceae), with special reference to population sizes . American Journal of Botany , 90 : 897 – 904 . doi: 10.3732/ajb.90.6.897