ABSTRACT
Common vetch (Vicia sativa L.) is an important forage legume crop. Our study was focused on estimation of genotypic variation for seed traits among germplasm accessions within the V. sativa and the two subspecies of common vetch; V. sativa (402 accessions), V. sativa subsp. sativa (105 accessions) and V. sativa subsp. nigra (30 accessions). The seed traits measured were straight length, straight width, width to length ratio, curved length, curved width, perimeter, hilum length, 100-seed weight and seed shape. The seed trait data were analysed using REML in GenStat and the resulting accession-by-trait BLUP mean matrices were summarised using a combination of cluster and principal component analysis, presented as biplots. There was significant (P < 0.05) genotypic variation among germplasm accessions, within each subspecies for all the traits measured. The calculated seed trait repeatability (R) provided a rough estimate of the upper limit of genotypic variation among the accessions within the V. sativa and the two subspecies. The magnitude and type of association among the seed traits shown in the biplots were supported by the estimated phenotypic correlation coefficients. The germplasm accessions within the groups identified in V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra will provide valuable genetic diversity for taxonomic studies and breeding associated with the seed morphological traits reported in our investigation.
Introduction
Common vetch (Vicia sativa L.) is an important self-pollinating annual forage legume (Chooi Citation1971; Firincioglu et al. Citation2009; Chung et al. Citation2013; Liu et al. Citation2013; Mikić et al. Citation2013). It is used as a cover crop, green manure, pasture, and for silage and hay production (Sullivan & Diver Citation2003; Kahlaoui et al. Citation2009; Yolcu Citation2011; Firincioglu Citation2012; Kim et al. Citation2015). Common vetch also grows well in mixtures with field crops and provides cool season forage. It has the ability to fix atmospheric nitrogen and grow across a wide range of climates and soil types (Chung et al. Citation2014; Liu et al. Citation2014; Kim et al. Citation2015). Due to its multiple uses and broad environmental adaptation, it is widely grown in Turkey, China and other parts of the world (Wang & Ren Citation1989; Arslan et al. Citation2012). In Turkey, V. sativa has the largest cultivated area of 579,684 ha compare with other annual feed legumes (Firincioglu Citation2012).
Based on phenotypic traits, Maxted (Citation1993) subdivided the subgenus Vicia L. into nine series, 38 species, 14 subspecies and 22 varieties, and confirmed that this genus was mainly distributed in the temperate regions of the northern hemisphere and Latin America. The Chinese Virtual Herbarium (CVH) (http://www.cvh.org.cn/) and Song et al. (Citation2011) reported that this species is also widely distributed in China.
Germplasm is considered as the genetic raw material used to develop new and superior crop cultivars (Fehr Citation1987). The genetic diversity present in common vetch germplasm will therefore provide the genetic base for cultivar development. The efficient utilisation of the genetic variation present in a germplasm collection depends on the availability of information on the various traits present in the accession (Beuselinck & Steiner Citation1992). Characterisation of germplasm collections will provide useful information for plant breeding programmes (McIvor & Bray Citation1983). Studies of the variation present in germplasm collections have been carried out on many plant species for a wide range of traits or attributes. Characterisation of plant morphological attributes has been reported in durum wheat (Triticum turgidum L. conv. durum) (Pecetti et al. Citation1992), potato (Solanum tuberosum L.) (Huaman Citation1984), tall fescue (Veronesi & Falcinelli Citation1988), alfalfa (Rumbaugh et al. Citation1988), red clover (Kouame & Quesenberry Citation1993) and white clover (Jahufer et al. Citation1997).
The range of structural diversity available for seed traits is useful in the characterisation of variation within and among species (Barthlott Citation1981; Ocampo et al. Citation2014). The use of seed traits to characterise genetic diversity in germplasm can be an advantage in comparison with other plant organs as seeds are easy to collect and store (Grillo et al. Citation2010). Seed morphological traits can be utilised for species identification as well as selection criteria in crop improvement programmes (Balkaya et al. Citation2009; Al-Ghamdi & Al-Zahrani Citation2010). Characterisation based on seed traits was used to assess the variation among 21 clones of Hevea brasiliensis and provided data for clone breeding (Omokhafe & Alika Citation2004). Seed traits were used to study the genotypic variation among 160 populations of winter squash germplasm collected from different sites in Turkey (Balkaya et al. Citation2009). Seed trait variation was also evaluated to determine taxonomic values for species identification (Fagúndez & Izco Citation2004; Davitashvili & Karrer Citation2010). The importance of macro and micro seed trait studies in quantifying genetic diversity and plant taxonomy was demonstrated by Grillo et al. (Citation2010).
This paper reports on a study of genotypic variation for seed morphological traits among a set of 537 common vetch germplasm accessions. Our study was focused on estimation of genotypic variation for seed traits within the V. sativa and the two subspecies: V. sativa subsp. sativa and V. sativa subsp. nigra. These species and subspecies are widely distributed in China and other parts of the world, sown across large areas (Kovářová et al. Citation2007; Castiglione et al. Citation2011; Song et al. Citation2011; Arslan et al. Citation2012; Firincioglu Citation2012). The potential for identifying germplasm accessions with useful seed trait combinations, for application in plant breeding programmes, was also investigated.
Materials and methods
Germplasm
A total of 537 germplasm accessions were evaluated in this study. This consisted of 402 V. sativa, 105 V. sativa subsp. sativa and 30 V. sativa subsp. nigra. The accessions were acquired from the National Plant Germplasm System of the United States.
Field trial
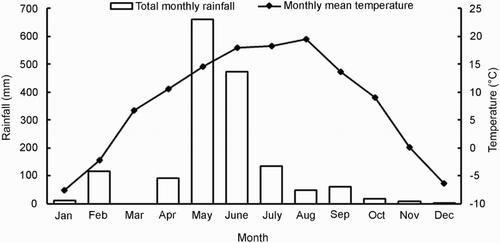
Seed of all 537 accessions were sown in a field experiment at Yuzhong (104°09′E, 35°89′N; 1720 m above sea level) in Gansu Province, China. At Yuzhong the soil type is loess. The mean monthly temperatures and total monthly rainfall during the trial period are shown in .
A week prior to planting, the soil was prepared as a fine seed bed to enhance good seedling establishment. The trial was planted on 1 May 2013. The field experimental layout was a randomised complete block design with three replicates. In each replicate, every accession was represented by a random sample of 10 seeds, planted in a 1 m row with a spacing of 10 cm between plants. The trial was managed according to standard local recommendations. Irrigation was not applied during the trial.
The accessions were harvested for seed during the period July to November 2013, depending on their maturity. The harvested seed was dried down to a moisture content of between 9% and 10% prior to storage at 5 °C. Each accession was represented by three random samples of 50 seeds each. Each sample represented the row of plants harvested per replicate.
Trait measurements
A total of nine seed traits were measured in this study: SL, straight length (mm); SW, straight width (mm); WL, width to length ratio; CL, curved length (mm); CW, curved width (mm); Pe, perimeter (mm); HL, hilum length (mm); SY, 100-seed weight (g) and shape (circular/square, oval, cone and irregular) (Figure S1).
In order to conduct the trait measurements, images of seed samples were first obtained using a flatbed scanner (EPSON GT-15000). The scanning was conducted using an image resolution of 200 dpi and a scanning area of more than 1024 × 1024 pixels. A room temperature of 20 °C and 15% relative humidity was maintained during scanning to avoid any possible variation in seed size, weight, shape and colour (Venora et al. Citation2007, Citation2009; Grillo et al. Citation2010).
For each accession, scanning was conducted on each of the three replicate samples, consisting of 50 seeds. Seed sample preparation and scanning were conducted according to methods used by Venora et al. (Citation2007, Citation2009), Bacchetta et al. (Citation2008), Balkaya et al. (Citation2009) and Grillo et al. (Citation2010).
Image analysis
The WinSEEDLE 2011 image analysis system was used for image processing. WinSEEDLE is an image analysis system designed for narrow needle-shaped leaves, seed morphology and disease analysis. To digitise needles or seeds, WinSEEDLE uses an optical scanner with a special lighting system instead of a video camera. The scanner produces high resolution images free of illumination error. It automatically detects and analyses needles and seeds much more precisely than conventional area meters.
Data analysis
Data analysis was based on: (1) the variance component analysis procured to assess the significance and magnitude of genotypic variation among accessions within a species; and (2) pattern analysis, which consisted of a combination of cluster and principal component analysis (PCA) (Gabriel Citation1971; Kroonenberg Citation1994; Watson et al. Citation1995) to provide a graphical summary of the accession-by-multi trait data matrices.
The variance component analysis was conducted using the Residual Maximum Likelihood (REML) option, in GenStat 7.1 (2003). A random linear model was used to analyse data within subspecies data. The analysis generated accession means, for the seed traits, based on Best Linear Unbiased Predictors (BLUP) (White & Hodge Citation1989). A mixed linear model analysis with subspecies as a fixed effect was also carried out to investigate differences among V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra for each of the traits measured.
Accession mean repeatability (R) was estimated for each trait, according to Fehr (Citation1987), using the genotypic () and experimental error (
) variance components from REML analysis together with nr (number of replications):
(1) Phenotypic correlation (rp) analysis among the seed traits was carried out using GenStat 7.1 (2003).
Results
There was significant (P < 0.05) genotypic variation among the germplasm accessions within the V. sativa and the two subspecies,V. sativa subsp. sativa and V. sativa subsp. nigra, for all the seed traits measured at Yuzhong (). This was also shown by the accession mean repeatability (R) estimates for the different traits within the V. sativa and the two subspecies. V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra had similar R estimates for the traits CL, CW, Pe and SW. Trait SW had the highest R value in V. sativa and the two subspecies. The R estimates for the traits SY, HL and SL were low within the V. sativa and the two subspecies. The width to length ratio had the lowest R estimate (). Analysis of average individual trait expression across V. sativa and the two subspecies indicated significant (P < 0.05) differences for all the eight traits measured ().
Table 1. Average, maximum, minimum, least significant differences (LSD0.05), estimated genotypic ( ) and experimental error (
) and experimental error ( ) variance components and associated standard error (±SD), among the 402, 105 and 30 germplasm accessions of V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra, respectively, evaluated at Yuzhong.
) variance components and associated standard error (±SD), among the 402, 105 and 30 germplasm accessions of V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra, respectively, evaluated at Yuzhong.
The variation for seed shape within V. sativa and the two subspecies was relatively low. All of the accessions except for seven in V. sativa had circular- and square-shaped seeds. V. sativa had one cone-shaped, two irregular-shaped and four oval-shaped accessions (Figure S1).
Pattern analysis and phenotypic correlation
Principal component analysis (PCA) of the accession-by-trait BLUP adjusted mean matrices generated biplots (–) for all subspecies. The correlation structure among the traits in each biplot is indicated by the directional vectors. The symbols in each biplot indicated the groupings generated from cluster analysis.
Figure 2. Biplot generated using standardised Best Linear Unbiased Predictor values for eight traits measured from the 402 germplasm accessions of V. sativa at Yuzhong. Components I and II account for 71% and 14% of total variation, respectively. The different symbols indicate accession groups 1 to 5 generated from cluster analysis. Traits are indicated by the directional vectors: SY, 100-seed weight; CL, curved length; CW, curved width; HL, hilum length; Pe, perimeter (mm); SL, straight length (mm); SW, straight width (mm); WL, width to length ratio.
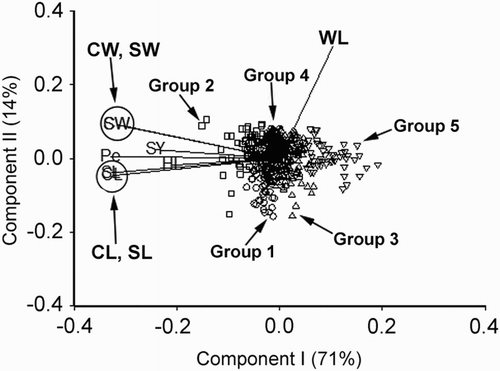
Figure 3. Biplot generated using standardised Best Linear Unbiased Predictor values for eight traits measured from the 105 germplasm accessions of V. sativa subsp. sativa at Yuzhong. Components I and II account for 74% and 13% of total variation, respectively. The different symbols indicate accession groups 1 to 4 generated from cluster analysis. Traits are indicated by the directional vectors: SY, 100-seed weight; CL, curved length; CW, curved width; HL, hilum length; Pe, perimeter (mm); SL, straight length (mm); SW, straight width (mm); WL, width to length ratio.
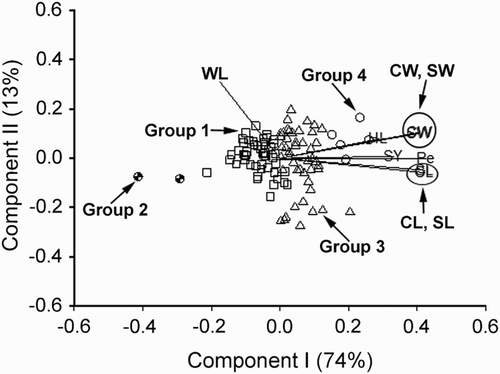
Figure 4. Biplot generated using standardised Best Linear Unbiased Predictor values for eight traits measured from the 30 germplasm accessions of V. sativa subsp. nigra at Yuzhong. Components I and II account for 81% and 9% of total variation, respectively. The different symbols indicate accession groups 1 to 3 generated from cluster analysis. Traits are indicated by the directional vectors: SY, 100-seed weight; CL, curved length; CW, curved width; HL, hilum length; Pe, perimeter (mm); SL, straight length (mm); SW, straight width (mm); WL, width to length ratio.
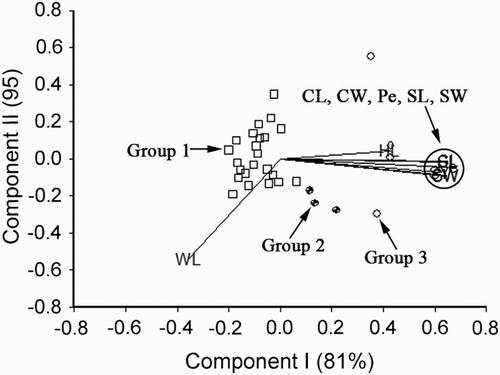
V. sativa
The traits SY, CL, CW, HL, Pe, SL and SW showed a strong positive association (angles between the directional vectors are at <90°) (). Trait WL was negatively associated (angles between the directional vectors are at >90°) with these traits. Cluster analysis of the 402 germplasm accessions based on the eight traits SY, CL, CW, HL, Pe, SL, SW and WL, generated five groups. Group 2 contained accessions with above-average expression for the traits SY, CL, CW, HL, Pe, SL and SW. The members of groups 5 and 3 had below-average expression for these traits.
summarises the characteristics of the five V. sativa accession groups generated from cluster analysis based on the traits SY, CL, CW, HL, Pe, SL, SW and WL. Accessions in group 5 had the lowest expression for these traits. This group was also the smallest group consisting of 48 accessions in contrast to group 3 with 145 accessions.
Table 2. Trait means for each of the five groups of the 402 V. sativa germplasm accessions, generated from pattern analysis. Group number and members per group are indicated.
The phenotypic correlation coefficients (rp) shown in support the association among the traits SY, CL, CW, HL, Pe, SL, SW and WL indicated in . Positive rp ranged from a moderate 0.498 between HL and SW to strong 0.999 between SW and CW.
Table 3. Phenotypic (rp) correlation coefficients, between traits based on the 402 V. sativa germplasm accessions evaluated at Yuzhong.
V. sativa subsp. sativa
The traits SY, CL, CW, HL, Pe, SL and SW had a strong positive association (angles between the directional vectors are at <90°) among each other within the set of 105 germplasm accessions (). WL was negatively associated with all the other traits. The four groups generated from cluster analysis of the 105 accessions of V. sativa subsp. sativa, based on the accession-by-trait matrix, indicated by the different symbols in showed a relatively good separation among the groups. Group 4 clearly contained accessions with above-average expression for the traits SY, CL, CW, HL, Pe, SL and SW. Some of the accessions within group 3 had above-average expression for the traits WL, HL, CW and SW.
Group means for the eight traits SY, CL, CW, HL, Pe, SL, SW and WL are presented in . Group 4 with five accession members had the highest mean expression for all traits except WL. Group 2 containing two accessions had the lowest mean expression for all traits, except for WL.
Table 4. Trait means for each of the four groups of the 105 V. sativa subsp. sativa germplasm accessions, generated from pattern analysis. Group number and members per group are indicated.
These relationships among traits SY, CL, CW, HL, Pe, SL, SW and WL, shown in , were confirmed by the estimated phenotypic correlation coefficients (rp) in . While the traits CW and SW had the strongest positive rp, the traits CL and HL had the lowest positive rp. The negative association of trait WL with the other traits, shown in the biplot () was also confirmed by the estimates of rp in .
Table 5. Phenotypic (rp) correlation coefficients, between traits based on the 105 V. sativa subsp. sativa germplasm accessions evaluated at Yuzhong.
V. sativa subsp. nigra
Principal component analysis of the 30 germplasm accessions generated a bilpot () showing a strong positive association among the traits SY, CL, CW, HL, Pe, SL and SW. The trait WL was negatively associated with SY, CL, CW, HL, Pe, SL and SW. Cluster analysis of the 30 accessions based on their expression of the traits SY, CL, CW, HL, Pe, SL, SW and WL resulted in generating three groups, indicated by the different symbols (). These groups were clearly differentiated with no overlap. Group 3 had above-average expression for all traits except WL. Group 2 also consisted of accessions with above-average expression for the traits SY, CL, CW, HL, Pe, SL and SW, but were not as high in expression as the members in group 3. Most of the accessions in group 1 were well below average expression for all the traits, except WL.
The group averages for the traits SY, CL, CW, HL, Pe, SL, SW and WL are presented in . Group 3 had the highest means for all the traits except WL. Group 1 had the lowest trait means. These results were graphically summarised in the biplot in . Group 1, the largest, consisted of 23 accessions.
Table 6. Trait means for each of the three groups of the 30 V. sativa subsp. nigra germplasm accessions, generated from pattern analysis. Group number and members per group are indicated.
Phenotypic correlation coefficients (rp) estimated in , from the accession-by-trait matrix, confirms the association among the traits SY, CL, CW, HL, Pe, SL, SW and WL indicated in . There were strong positive estimates of rp for all the pairwise combinations of the traits SY, CL, CW, Pe, SL and SW. As indicated in , the rp estimates for HL and the traits SY, CL, CW, Pe, SL and SW were positive and moderate (). The estimates of rp of WL with SY, CL, CW, Pe, SL and SW were negative and moderate except for HL which was also negative but low.
Table 7. Phenotypic (rp) correlation coefficients, between traits based on the 30 V. sativa subsp. nigra germplasm accessions evaluated at Yuzhong.
Discussion
In our study, there was significant (P < 0.05) genotypic variation among germplasm accessions, within V. sativa and the two subspecies, for all the measured seed traits: SY, CL, CW, HL, Pe, SL, SW and WL. The estimates of genotypic variation and repeatability for these traits indicated the potential genetic variation available among the germplasm accessions within V. sativa and the two subspecies investigated. With regard to some of the individual traits, SY ranged from 0.44–5.39 g, 0.75–5.01 g and 0.48–3.64 g among the V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra accessions, respectively. Seed length ranged from 2.25–6.35 mm, 2.37–6.18 mm and 2.18–5.53 mm for V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra, respectively. The trait SW among the accessions of the V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra, ranged from 2.41–5.94 mm, 2.28–5.59 mm and 2.1–5.1 mm, respectively.
These traits associated with seed size are important for seed establishment and survival. According to Aarssen & Jordan (Citation2001), Henery & Westoby (Citation2001) and Moles et al. (Citation2005), small-seeded species are able to produce more seeds for a given amount of energy in comparison with large-seeded species. However, large-seeded species develop seedlings that are able to tolerate many of the stresses encountered during their establishment. This indicates that seed size of different species growing in a particular environment may have a significant impact on seedling establishment and survival. Therefore, information on the magnitude of genetic variation for key seed size traits among germplasm accessions will enhance cultivar development programmes (Odiaka Citation2005), focused on improving seed yield and seedling establishment/survival. The position of each of the germplasm groups relative to the directional vectors enabled identification of accessions potentially useful for improving seed size. At present, common vetch is mainly planted in the Qinghai-Tibetan plateau of China. But this area has a short growing season and harsh environmental conditions (Nan et al. Citation2006; Mao et al. Citation2015). Therefore, it is better to use large seeds to improve seedling establishment and survival.
In our reported study, in addition to the significant (P < 0.05) genotypic variation for the different seed traits estimated within each subspecies, comparison of individual trait averages among V. sativa and the two subspecies indicated significant (P < 0.05) differences. These significant (P < 0.05) differences in trait expression among V. sativa and the two subspecies, could be useful in investigating their taxonomic relationships (Agulló et al. Citation1991). Grillo et al. (Citation2010), Bécquer et al. (Citation2014) and Ocamp et al. (Citation2014) have used digital images of seeds to study genetic diversity and species classification. These digital images include information such as variation in length, width, perimeter and also seed characteristics such as shape. These results indicate the potential of using database information on seed morphology for taxonomic screening (Dell’Aquila Citation2007).
The application of principal component and cluster analysis to the germplasm accession-by-seed trait BLUP adjusted mean matrices, of each of the species and subspecies V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra, provided a graphical summary of the genotypic variation and trait association. The type and magnitude of association among the directional vectors in the biplots, representing the different traits, was supported by the estimated rp for all the species and subspecies. The association of the eight traits were similar in all the species and subspecies, where SY, CL, CW, HL, Pe, SL and SW were positively correlated. The trait WL was negatively correlated with all the seven other traits.
The use of pattern analysis to identify elite material in breeding programmes has been reported by: Harch et al. (Citation1996) for groundnut (Arachis hypogaea L.), Jahufer et al. (Citation1997) for white clover (Trifolium repens L.) and Zhang et al. (Citation2006) for spring wheat (Triticum aestivum). Pattern analysis carried out within each of the species and subspecies V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra enabled identification of accession groups with potential to be used for improvement of the seed traits examined in our study. The germplasm accessions within the groups identified in V. sativa, V. sativa subsp. sativa and V. sativa subsp. nigra will provide valuable genetic diversity for taxonomic studies and breeding associated with the seed morphological traits reported in our investigation.
Figure S1. Diagram of seed shape trait measurement.
Download TIFF Image (1.5 MB)Acknowledgements
The authors express their sincere thanks to the US National Plant Germplasm System (NPGS) for providing the accessions used in the study. We also acknowledge Qiang Zhou, Dong Luo, Kai Luo, Rui Zhang, Lichao Ma, Yu Wang, Tianlong Chen and Peng Liu for their valuable help and advice.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aarssen LW, Jordan CY. 2001. Between-species patterns of covariation in plant size, seed size and fecundity in monocarpic herbs. Ecoscience. 8:471–477.
- Agulló MA, Tortosa ME, Ceresuela JL, Ortiz JM. 1991. Seed morphology in several taxa of the subfamily Papilionidae (Fabaceae). Botanika Chronika. 10:651–660.
- Al-Ghamdi FA, Al-Zahrani RM. 2010. Seed morphology of some species of Tephrosia pers. (Fabaceae) from Saudi Arabia identification of species and systematic significance. Am J Plant Sci. 121:59–65.
- Arslan E, Ertuğrul K, Öztürk AB. 2012. Karyological studies of some species of the genus Vicia L. (Leguminosae) in Turkey. Caryologia. 65:106–113. doi: 10.1080/00087114.2012.709804
- Bacchetta G, Grillo O, Mattana E, Venora G. 2008. Morpho-colorimetric characterisation by image analysis to identify diaspores of wild plant species. Flora. 203:669–682. doi: 10.1016/j.flora.2007.11.004
- Balkaya A, Yanmaz R, Özbakir M. 2009. Evaluation of variation in seed characters of Turkish winter squash (Cucurbita maxima) populations. New Zeal J Crop Hort. 37:167–178. doi: 10.1080/01140670909510262
- Barthlott W. 1981. Epidermal and seed surface characters of plants: systematic applicability and some evolutionary aspects. Nord J Bot. 1:345–355. doi: 10.1111/j.1756-1051.1981.tb00704.x
- Bécquer ER, Michelangeli FA, Borsch T. 2014. Comparative seed morphology of the Antillean genus Calycogonium (Melastomataceae: Miconieae) as a source of characters to untangle its complex taxonomy. Phytotaxa. 166:241–258. doi: 10.11646/phytotaxa.166.4.1
- Beuselinck PR, Steiner JJ. 1992. A proposed framework for identifying, quantifying, and utilising plant germplasm resources. Field Crops Res. 29:261–272. doi: 10.1016/0378-4290(92)90029-9
- Castiglione MR, Frediani M, Gelat MT, Ravalli C, Venor G, Caputo P, Cremonin R. 2011. Cytology of Vicia species. X. Karyotype evolution and phylogenetic implication in Vicia species of the sections Atossa, Microcarinae, Wiggersia and Vicia. Protoplasma. 248:707–716. doi: 10.1007/s00709-010-0232-7
- Chooi WY. 1971. Variation in nuclear DNA content in the genus Vicia. Genetics. 68:195–211.
- Chung J-W, Kim TS, Sundan S, Lee G-A, Park J-H, Cho G-T, Lee H-S, Lee J-Y, Lee M-C, Baek H-J, Lee SY. 2014. New cDNA-SSR markers in the narrow-leaved vetch (Vicia sativa subsp. nigra) using 454 pyrosequencing. Mol Breed. 33:749–754. doi: 10.1007/s11032-013-9980-3
- Chung J-W, Kim T-S, Suresh S, Lee S-Y, Cho G-T. 2013. Development of 65 novel polymorphic cDNA-SSR markers in common vetch (Vicia sativa subsp. sativa) using next generation sequencing. Molecules. 18:8376–8392. doi: 10.3390/molecules18078376
- Davitashvili N, Karrer G. 2010. Taxonomic importance of seed morphology in Gentiana (Gentianaceae). Bot J Linn Soc. 162:101–115. doi: 10.1111/j.1095-8339.2009.01020.x
- Dell'Aquila A. 2007. Towards new computer imaging techniques applied to seed quality testing and sorting. Seed Sci Technol. 35:519–538. doi: 10.15258/sst.2007.35.3.01
- Fagúndez J, Izco J. 2004. Taxonomic value of seed characters in the Erica tetralix L. group (Ericaceae). Plant Biosyst. 138:207–213. doi: 10.1080/11263500400021059
- Fehr WR. 1987. Principles of cultivar development. vol 1. New York: Macmillan publishing company.
- Firincioglu HK. 2012. A comparison of six vetches (Vicia spp.) for developmental rate, herbage yield and seed yield in semi-arid central Turkey. Grass Forage Sci. 69:303–314. doi: 10.1111/gfs.12021
- Firincioglu HK, Erbektas E, Dogruyol L, Mutlu Z, Ünal S, Karakurt E. 2009. Phenotypic variation of autumn and spring-sown vetch (Vicia sativa ssp.) populations in central Turkey. Span J Agric Res. 7:596–606. doi: 10.5424/sjar/2009073-444
- Gabriel KR. 1971. The biplot graphic display of matrices with application to principal component analysis. Biometrika. 58:453–467. doi: 10.1093/biomet/58.3.453
- Grillo O, Mattana E, Venora G, Bacchetta G. 2010. Statistical seed classifiers of 10 plant families representative of the Mediterranean vascular flora. Seed Sci Technol. 38:455–476. doi: 10.15258/sst.2010.38.2.19
- Harch BD, Basford KE, DeLacy IH, Lawrence PK, Cruickshank A. 1996. Mixed data types and the use of pattern analysis on the Australian groundnut germplasm data. Genet Resour Crop Evol. 43:363–376. doi: 10.1007/BF00132957
- Henery ML, Westoby M. 2001. Seed mass and seed nutrient content as predictors of seed output variation between species. Oikos. 92:479–490. doi: 10.1034/j.1600-0706.2001.920309.x
- Huaman Z. 1984. The evaluation of potato germplasm at the International Potato Centre. In: Holden JHW, Williams JT, editors. Crop genetic resources: conservation and evaluation. 1st ed. London: George Allen & Unwin; p. 199–201.
- Jahufer MZZ, Cooper M, Harch BD. 1997. Pattern analysis of the diversity of morphological plant attributes and herbage yield in a world collection of white clover (Trifolium repens L.) germplasm characterised in a summer moisture stress environment of Australia. Genet Resour Crop Evol. 44:289–300. doi: 10.1023/A:1008692629734
- Kahlaoui S, Walker DJ, Correal E, Martinez-Gomez P, Hassen H, Bouzid S. 2009. The morphology, chromosome number and nuclear DNA content of Tunisian populations of three Vicia species. Afr J Biotechnol. 8:3184–3191.
- Kim T-S, Raveendar S, Suresh S, Lee G-A, Lee J-R, Cho J-H, Lee S-Y, Ma K-H, Cho G-T, Chung J-W. 2015. Transcriptome analysis of two Vicia sativa subspecies: mining molecular markers to enhance genomic resources for vetch improvement. Genes. 6:1164–1182. doi: 10.3390/genes6041164
- Kouamé CN, Quesenberry KH. 1993. Cluster analysis of a world collection of red clover germplasm. Genet Resour Crop Evol. 40:39–47. doi: 10.1007/BF00053463
- Kovářová P, Navrátilová A, Macas J, Doležel J. 2007. Chromosome analysis and sorting in Vicia sativa, using flow cytometry. Biol Plantarum. 51:43–48. doi: 10.1007/s10535-007-0009-9
- Kroonenberg PM. 1994. The TUCKALS line: a suite of programs for three-way data analysis. Comput Stat Data An. 18:73–96. doi: 10.1016/0167-9473(94)90133-3
- Liu ZP, Liu P, Luo D, Liu WX, Wang YR. 2014. Exploiting illumina sequencing for the development of 95 novel polymorphic EST-SSR markers in common vetch (Vicia sativa subsp. sativa). Molecules. 19:5777–5789. doi: 10.3390/molecules19055777
- Liu ZP, Ma LC, Nan ZB, Wang YR. 2013. Comparative transcriptional profiling provides insights into the evolution and development of the zygomorphic flower of Vicia sativa (Papilionoideae). PloS One. 8:e57338. doi: 10.1371/journal.pone.0057338
- Mao ZX, Hua F, Nan ZB, Wan CG. 2015. Fatty acid, amino acid, and mineral composition of four common vetch seeds on Qinghai-Tibetan plateau. Food Chem. 171:13–18. doi: 10.1016/j.foodchem.2014.08.090
- Maxted N. 1993. A phenetic investigation of Vicia L. subgenus Vicia (Leguminosae, Vicieae). Bot J Linn Soc. 111:155–182. doi: 10.1111/j.1095-8339.1993.tb01897.x
- McIvor JG, Bray RA. 1983. Genetic resources of forage plants. Melbourne: CSIRO; p. 313–321.
- Mikić A, Mihailović V, Ćupina B, Vasiljević S, Milošević B, Katanski S, Matić R, Radojević V, Kraljević-Balalić, M. 2013. Agronomic characteristics related to grain yield and crude protein content in common vetch (Vicia sativa) accessions of diverse geographic origin. New Zeal J Agr Res. 56:297–308. doi: 10.1080/00288233.2013.845231
- Moles AT, Ackerly DD, Webb CO, Tweddle JC, Dickie JB, Westoby M. 2005. A brief history of seed size. Science. 307:576–580. doi: 10.1126/science.1104863
- Nan ZB, El-Moneim AMA, Larbi A, Nie B. 2006. Productivity of vetches (Vicia spp.) under alpine grassland conditions in China. Trop Grasslands. 40:177–182.
- Ocampo G, Michelangeli FA, Almeda F. 2014. Seed diversity in the tribe Miconieae (Melastomataceae): taxonomic, systematic, and evolutionary implications. PloS One. 9:e100561. doi: 10.1371/journal.pone.0100561
- Odiaka N. 2005. Morphological diversity among local germplazm of fluted pumpkin collected in Makurdi, Nigeria. J Food Agr Environ. 3:199–204.
- Omokhafe KO, Alika JE. 2004. Clonal variation and correlation of seed characters in Hevea brasiliensis muell. arg. Ind Crop Prod. 19:175–184. doi: 10.1016/j.indcrop.2003.09.004
- Pecetti L, Annicchiarico P, Damania AB. 1992. Biodiversity in a germplasm collection of durum wheat. Euphytica. 60:229–238.
- Rumbaugh MD, Graves WL, Caddel JL, Mohammad RM. 1988. Variability in a collection of alfalfa germplasm from Morocco. Crop Sci. 28:605–609. doi: 10.2135/cropsci1988.0011183X002800040005x
- Song M, Yu HZ, Lou YJ, Xu Ak. 2011. Biological characteristics and use of Vicia amoena. Prataculture Anim Husbandry. 4:5–6.
- Sullivan PG, Diver S. 2003. Overview of cover crops and green manures. Fayetteville: NCAT Agriculture Specialist.
- Venora G, Grillo O, Ravalli C, Cremonini R. 2009. Identification of Italian landraces of bean (Phaseolus vulgaris L.) using an image analysis system. Sci Hortic. 121:410–418. doi: 10.1016/j.scienta.2009.03.014
- Venora G, Grillo O, Shahin MA, Symons SJ. 2007. Identification of Sicilian landraces and Canadian cultivars of lentil using an image analysis system. Food Res Int. 40:161–166. doi: 10.1016/j.foodres.2006.09.001
- Veronesi F, Falcinelli M. 1988. Evaluation of an Italian germplasm collection of Festuca arundinacea Schreb. through a multivariate analysis. Euphytica. 38:211–220. doi: 10.1007/BF00023523
- Wang D, Ren JZ. 1989. Forage science. 2nd ed. Nanjing: Jiangsu Science and Technology Press; p. 191–202.
- Watson SL, DeLacy IH, Podlich DW, Basford KE. 1995. GEBEIL: ananalysis package using agglomerative hierarchical classificatory and SVD ordination procedures for genotype × environment data. Brisbane, Australia: Centre for Statistics Research Report, Dep. of Agriculture, Univ. of Queensland.
- White TL, Hodge GR. 1989. Predicting breeding values with applications in forest tree improvement. Forestry Sciences 33. Boston, MA: Kluwer Academic.
- Yolcu H. 2011. The effects of some organic and chemical fertilizer applications on yield, morphology, quality and mineral content of common vetch (Vicia sativa L.). Turk J Field Crops. 16:197–202.
- Zhang Y, He ZH, Zhang AM, Maarten VG, Pena RJ, Ye GY. 2006. Pattern analysis on protein properties of Chinese and CIMMYT spring wheat cultivars sown in China and CIMMYT. Aust J Agr Res. 57:811–822. doi: 10.1071/AR05372