Abstract
Polyploidy is common in the widespread genus Plantago, and might be especially important in the evolutionary history of native New Zealand species of the genus. To further understanding of native New Zealand Plantago, mitotic and meiotic chromosome counts are reported from 58 individuals representing most native species and subspecies, and one introduced species, complementing previous studies by extending the number of individuals and the geographic range of taxa counted. Previous counts were confirmed for most Plantago lanigera (2n = 2x = 12), all P. raoulii, P. spathulata subsp. spathulata, P. spathulata subsp. picta, P. triandra subsp. triandra and P. triandra subsp. masoniae (2n=8x=48), most P. unibracteata (2n=10x=60), and all P. sp. ‘Sylvester’ (2n=16x=96). Novel counts include 2n=12x=72 for four individuals identified as P. unibracteata, and 2n=48 for four individuals from Sugarloaf Pass, South Island, referred to here as P. aff. spathulata. One of the eight naturalized species, P. major, was diploid with 2n=12. These results, together with other studies, suggest that all native New Zealand Plantago have a base chromosome number of x=6 and most polyploids are allopolyploids.
Introduction
Approximately two-thirds of the>200 species of the cosmopolitan genus Plantago have had at least one chromosome count (Rahn Citation1996, who included Bougueria and Litorella, but see Hoggard et al. Citation2003). The basic chromosome number of Plantago is considered to be x=6, and the ancestor of the genus was probably diploid with 2n=12 (Rahn Citation1996). Roughly 75% of the studied species (representing all six subgenera) have x=6 as a base number, whereas the remaining 25% of Plantago species (from three subgenera) have x=5 (Rahn Citation1957, Citation1996). In a phylogenetic study based mostly on morphological characters, Rahn (Citation1996) inferred three independent reductions in Plantago from x=6 to x=5, as well as one reduction to x=4.
Previous cytological studies of New Zealand Plantago (all with a base number of x=6) reveal an intriguing evolutionary history in which polyploidy has clearly played an important role. The first reported counts of New Zealand Plantago were 2n=48 for P. spathulata (Rahn Citation1957), and n=24/2n=48 for P. raoulii, P. spathulata, and P. triandra, and 2n=24 for ‘P. brownii’ (Rattenbury Citation1957). (This ‘P. brownii’ is likely to be P. novae-zelandiae and was referred to as P. aff. lanigera by Dawson Citation2000). There was also a dubious count of 2n=18 for P. raoulii (McCullagh Citation1934; Dawson Citation2000). In these early studies, only one or very few individuals of each species were counted, and herbarium vouchers were not cited for many individuals. [The voucher for the P. raoulii accession counted by Rattenbury (Citation1957) is probably AK 46123, but vouchers for the other three individuals do not exist at AK (Ewen Cameron, personal communication).] Subsequently, Groves & Hair (Citation1971) greatly increased sampling of New Zealand Plantago to include counts from a total of 40 individuals of eight described species, all vouchered at CHR. They found 2n=12 for P. lanigera (five individuals), P. triantha (two individuals) and ‘P. novae-zelandiae diploid’ (six individuals); 2n=24 for ‘P. novae-zelandiae tetraploid’ (four individuals); 2n=48 for P. picta (one individual), P. raoulii (five individuals), P. spathulata (five individuals) and P. triandra (six individuals); and 2n=60 for P. unibracteata (as P. uniflora; five individuals). In addition, they found one individual from Lake Sylvester, NW Nelson to have n=48 and 2n=96, which they called P. sp. 16x (Groves & Hair Citation1971) and which was later dubbed P. sp. ‘Sylvester’ by Druce (Citation1993). More recently one individual of P. obconica was found to have 2n=12 (Dawson Citation1989).
Polyploidy in Plantago is also common on a global scale. Of the species of Plantago with chromosome counts, approximately two-thirds are polyploids. Nevertheless, polyploidy has been dismissed by some as playing only a minor role in shaping diversification for most of the genus (Briggs Citation1973; Rahn Citation1996), with the important exception of sect. Oliganthos from the large subgenus Plantago (Rahn Citation1984). By contrast, Ishikawa et al. (Citation2009) hypothesized that the entire subgenus Plantago has had a history of allopolyploidy both among and within sections. Indeed, of the 24 species of Plantago that are known to be hexaploid (2n=6x=36) or higher, 20 are from the subgenus Plantago, including nine from sect. Oliganthos and four from sect. Mesembrynia. For the most part, these two sections comprise species from the southern hemisphere, including all but one New Zealand native species (P. aucklandica, sect. Plantago) (Rahn Citation1996). When chromosome number was mapped onto a recent molecular phylogeny of Australasian species of these sections, it was shown that polyploid series have evolved at least four times in the Australasian group (Tay et al. Citation2010a). Tay et al. (Citation2010a) also concluded that sections as currently circumscribed are not all monophyletic. We believe that detailed cytological studies will play a key role in resolving the historically difficult taxonomy of the New Zealand species of Plantago, as was the case with their southern South American counterparts (Rahn Citation1984). With this aim in mind, we publish here the chromosome counts for accessions of most native species of New Zealand Plantago and one naturalized species.
Materials and methods
The individual plants used in this study were collected in the field, and the identifications and places of collection are listed in . When several individuals of a particular species of Plantago were collected at the same locality, they were given the same collector's number with a different letter (e.g. 250X, 250Y). One of these individuals was pressed to make a voucher for the population and these have been deposited either in WELT or WELTU (). The other plants have been kept alive and transplanted to greenhouses for this study and for further molecular cytogenetic research. A photograph of each plant included in this study was also taken, and attached to the voucher specimen together with a record of its chromosome number. When our molecular cytogenetic studies are completed the entire plant will be vouchered in WELT.
Table 1 The species, chromosome numbers, collection details and herbarium vouchers of the Plantago accessions used in this study.
Mitotic chromosomes were observed in root tips that were pretreated with a saturated solution of paradichlorobenzene for ~ 18 h at 4 °C and fixed in ethanol/acetic acid (3:1, v/v). The roots were hydrolysed in 1 M HCl at 60 °C for 6–8 min, transferred to 45% acetic acid and then macerated on a slide in a drop of FLP orcein (Jackson Citation1973), heated over a spirit flame, flattened under a coverslip and observed under the microscope. Meiotic chromosomes were obtained from immature anthers that were fixed in ethanol/chloroform/acetic acid (6:3:1) and stained with FLP orcein as above.
Results and discussion
Chromosome numbers were determined for 58 individuals representing most species and subspecies of native New Zealand Plantago and one introduced species (). Our sampling of these species complements that of Groves & Hair (Citation1971), which means the chromosome counts now more fully represent their geographical ranges on the North and South Islands. Our results confirm previous counts for most of the species, but we have found potentially new numbers for two species, including dodecaploid (2n=12x=72) for P. unibracteata, which is the first time this chromosome number has been recorded from New Zealand material.
All the plants of P. raoulii, P. spathulata subsp. spathulata, P. spathulata subsp. picta, P. triandra subsp. triandra and P. triandra subsp. masoniae were octoploid (2n=8x=48), as in previous studies.
Confirming previous reports, most plants of P. lanigera sampled were diploid (2n=2x=12). Octoploid (2n=8x=48) plants of individuals identified in the field as P. lanigera by the collector (Michael J. Thorsen, Department of Conservation) were counted from four individuals from Sugarloaf Pass (A). The leaf morphology of these plants indeed approaches P. lanigera, and yet we are not completely satisfied with this identification. Also, because these individuals cluster closely with P. spathulata and P. raoulii, and not P. lanigera, using ITS sequences (Tay et al. Citation2010b) and AFLP markers (Heidi M Meudt Citationin press), we refer to them here as ‘P. aff. spathulata?’ pending further morphlogical study. No tetraploid individuals were found in our sampling of P. lanigera plants. Tetraploids (2n=4x=24) have been previously reported for P. novae-zelandiae (sometimes considered to be a synonym of P. lanigera, and also called P. aff. lanigera by Dawson Citation2000) by Groves & Hair (Citation1971) from both Canterbury and Fiordland and by Rattenbury (Citation1957) from Otago. Plantago lanigera/P. novae-zelandiae thus appears to be variable chromosomally and more intensive sampling is needed to determine the geographical extent of its chromosome races.
Fig. 1 Somatic chromosomes of Plantago species. (A) Plantago aff. spathulata? (MLT049W; WELTU 20133) 2n=48; B, Plantago unibracteata (MLT 024W; WELTU 20175) 2n=72; C, Plantago sp. ‘Sylvester’ (BVS s.n. C; WELTU 20215) 2n=96. Scale bar=10 µm.
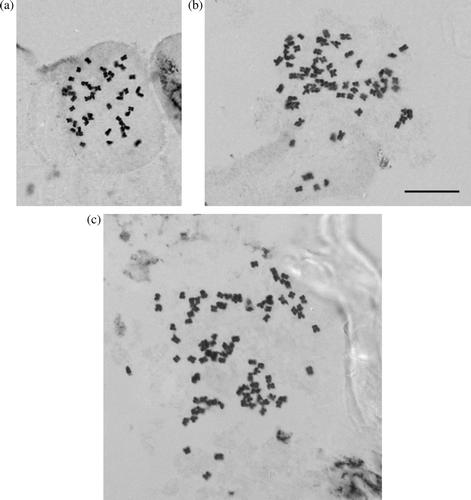
Most plants of P. unibracteata that we sampled were decaploid (2n=10x=60), but four Nelson individuals were found to be dodecaploid (2n=12x=72), including two individuals identified as P. unibracteata from Lake Sylvester, as well as one individual from Tableland near Balloon Hut (B). This third individual was originally identified as P. sp ‘Sylvester’ by the collector, but we identified it as P. unibracteata based on morphology, and AFLP markers clearly place it with other P. unibracteata and not with P. sp. ‘Sylvester’ (Heidi M Meudt Citationin press). No New Zealand species were known previously to have 2n=72, which has been found in only four other Plantago species worldwide (three South American species from sect. Oliganthos, and the European P. altissima in subg. Albicans; Rahn Citation1957, Citation1984). Whether these New Zealand individuals represent another chromosomal race of P. unibracteata, or a new alloploid species, involving an octoploid (2n=8x=48) and the 16-ploid (2n=16x=96) or perhaps more likely P. unibracteata (2n=10x=60) and a diploid species (2n=2x=12), requires further investigation.
Plants of the putative undescribed species P. sp. ‘Sylvester’ (Druce Citation1993) were verified to have 2n=16x=96 (C). Groves & Hair (Citation1971) counted one individual from Lake Sylvester, and here we report counts of seven additional individuals from three other localities, i.e. Boulder Lake, Tableland near Balloon Hut and Little Lake Sylvester. These data confirm the presence of this entity from several places in north-west Nelson. Only one other species of Plantago is known to share the 2n=96 count, P. correae (sect. Oliganthos) from southern South America (Rahn Citation1984).
Finally, we report 2n=12 for a naturalized plant from which Tay et al. (Citation2010a) previously obtained an ITS DNA sequence (as ‘P. sp.’; Genbank number FJ024620). This sequence is identical to another Genbank sequence (AY101862) from an individual identified as P. asiatica. The sequence is also 99% similar (1–3 nucleotide differences) to other Genbank sequences of individuals identified as P. asiatica (AJ548977), P. major (AY101861, FJ024619) and P. hostifolia (AB281166). Our count of 2n=12 matches all previous chromosome counts of P. major, but not those of P. asiatica (2n=24, Rahn Citation1957, Citation1996). The plants we counted fall within the published morphological range of P. major but differ from many New Zealand collections in having narrower, five-veined, cuneate leaves and shorter flowering portions to their spikes. For these reasons, we have identified the plants we examined as P. major rather than P. asiatica. It is possible that these represent a separate introduction of this widespread and variable species.
Meiosis was observed in three octoploid plants (P. aff. spathulata?, MLT 049Y and MLT 049Z; P. spathulata subsp. spathulata, PGJ 2629X) and in all cases chromosomes formed 24 bivalents at metaphase I. Bivalent formation was also reported by Groves & Hair (Citation1971) for octoploid P. spathulata and 16-ploid P. sp. ‘Sylvester’, and together with the results from Ishikawa et al. (Citation2009) this meiotic chromosome pairing behaviour would suggest that many, if not all, of the New Zealand polyploid species of Plantago are allopolyploids. Indeed, the identity and nature of all New Zealand Plantago, but especially P. sp. ‘Sylvester’ and the higher ploidy individuals of P. lanigera and P. unibracteata mentioned above, are being explored further with DNA sequencing (Tay et al. Citation2010b), molecular cytogenetics (C Wong & BG Murray unpublished observations), morphometrics (HM Meudt in press), and AFLP DNA fingerprinting (HM Meudt unpublished observations.
Acknowledgements
We thank Lesley Milicich for assistance with maintaining live plants in the Victoria University greenhouses and Dirk Albach for taxonomic advice. We also thank the Department of Conservation (especially Graeme Atkins, John Barkla, and Mike Thorsen), Arnold Dench, David Glenny, Pete Lockhart, Bill and Nancy Malcolm, Jessie Prebble and Barry Sneddon for assistance with field collections. This research was funded in part by the New Zealand Foundation for Research Science and Technology through the Defining New Zealand's Land Biota OBI.
References
- Briggs , BG . 1973 . Chromosomal studies on Plantago in Australia . Contributions from the NSW National Herbarium , 4 : 399 – 405 .
- Dawson , MI . 1989 . Contributions to a chromosome atlas of the New Zealand flora – 30. Miscellaneous species . New Zealand Journal of Botany , 27 : 163 – 165 .
- Dawson , MI . 2000 . Index of chromosome numbers of indigenous NewZealand spermatophytes . New Zealand Journal of Botany , 38 : 47 – 150 .
- Druce AP 1993 . Indigenous vascular plants of New Zealand, 9th revision . Landcare Research, Lower Hutt. Unpublished list held at WELT .
- Groves , BE and Hair , JB . 1971 . Contributions to a chromosome atlas of the New Zealand flora – 15. Miscellaneous families . New Zealand Journal of Botany , 9 : 569 – 575 .
- Hoggard , RK , Kores , PJ , Molvray , M , Hoggard , GD and Broughton , DA . 2003 . Molecular systematics and biogeography of the amphibious genus Littorella (Plantaginaceae) . American Journal of Botany , 90 : 429 – 435 .
- Ishikawa , N , Yokoyama , J and Tsukaya , H . 2009 . Molecular evidence of reticulate evolution in the subgenus Plantago (Plantaginaceae) . American Journal of Botany , 96 : 1627 – 1635 .
- Jackson , RC . 1973 . Chromosome evolution in Haplopappus gracilis: a centric transposition race . Evolution , 27 : 243 – 256 .
- McCullagh , D . 1934 . Chromosome and chromosome morphology in Plantaginaceae I . Genetica , 16 : 1 – 44 .
- Meudt HM in press . AFLP data reveal a history of auto- and allopolyploidy in the NewZealand endemic species of Plantago (Plantaginaceae): Newperspectives on a taxonomically – challenging group . International Journal of Plant Science .
- Rahn , K . 1957 . Chromosome numbers in Plantago . Botanisk Tidsskrift , 53 : 369 – 378 .
- Rahn , K . 1984 . Plantago sect. Oliganthos in southern South America, a taxonomic revision . Nordic Journal of Botany , 4 : 601 – 627 .
- Rahn , K . 1996 . A phylogenetic study of the Plantaginaceae . Botanical Journal of the Linnean Society , 120 : 145 – 198 .
- Rattenbury , JA . 1957 . Chromosome numbers in New Zealand angiosperms . Transactions of the Royal Society of New Zealand , 84 : 936 – 938 .
- Tay , ML , Meudt , HM , Garnock-Jones , PJ and Ritchie , PA . 2010a . DNA sequences from three genomes reveal multiple long-distance dispersals and non-monophyly of sections in Australasian Plantago (Plantaginaceae) . Australian Systematic Botany , 23 : 47 – 68 .
- Tay ML , Meudt HM , Garnock-Jones PJ , Ritchie PA 2010b . Testing species limits of New Zealand Plantago (Plantaginaceae) using internal transcribed spacer (ITS) DNA sequences . New Zealand Journal of Botany 48 : 205 224 .