Abstract
As traditionally circumscribed, Simplicia buchananii is an endemic of north-west Nelson, New Zealand, with all known records coming from the Kahurangi National Park. Simplicia laxa is more widespread with historical records from the eastern Wairarapa, and Otago. Neither grass is common and both are listed as Threatened. In 2005, Simplicia was discovered near Taihape in the central North Island. These plants had morphological attributes of both S. buchananii and S. laxa. We explored genetic variation in Simplicia using amplified fragment length polymorphism DNA fingerprinting and DNA sequencing of the plastid trnL intron and trnL–F intergenic spacer and the nuclear ribosomal internal transcribed spacer and external transcribed spacer regions. Populations in the Taihape area are referable to S. laxa. However, they and plants from a site in North Otago are genetically distinct from other South Island S. laxa plants and their taxonomic status needs further exploration.
Introduction
The New Zealand endemic genus Simplicia (Poaceae: Poeae) was established by Kirk (Citation1897) based on specimens he had gathered in 1880 from Dry River Station (Ruamahanga Valley, eastern Wairarapa, North Island) along with those collected by Donald Petrie about 1880 from Waikouaiti, on the north-eastern Otago coastline, South Island, and which he named as Simplicia laxa Kirk. The genus is well differentiated, possessing one-flowered spikelets, minute unequal glumes and conspicuous lemma equal in length to the inconspicuous palea (Kirk Citation1897; Zotov Citation1971; Edgar & Connor Citation2000). A second species, Simplicia buchananii Zotov, was initially described as Poa uniflora by Buchanan (Citation1880) and was based on a gathering from Mt Arthur, north-west Nelson. The possession of one-flowered spikelets, reduced glumes and lemma equal to the palea placed this species within Simplicia, where it was initially treated as a variety of S. laxa (Zotov Citation1943), and then as a species (Zotov Citation1971). At the time of Zotov's Citation1971 treatment of the genus, and aside from the type collection, S. buchananii was known from only a few 1960s gatherings made from Gouland Downs, Mt Peel and the Cobb Valley.
Since its formal description in 1897, S. laxa has remained an enigmatic species scarcely known to New Zealand botanists. For his treatment of the genus, Zotov (Citation1971) was only able to work with the historical Kirk and Petrie specimens and a single live plant that had beengathered in 1969 from Castle Rock in the Old Man Range (Otago).
Because both species of Simplicia were evidently extremely uncommon they were among the first indigenous grasses to be listed as ‘Rare and Endangered’ (Given Citation1976). S. laxa has been listed under the various threat classification systems used over the last two decades as ‘Insufficiently Known’, ‘Endangered’, ‘Nationally Endangered’, and currently as ‘Nationally Critical’ (Given Citation1981; Williams & Given Citation1981, Citation1989; Given Citation1990; Cameron et al. Citation1993, Citation1995; de Lange et al. Citation1999, Citation2004, Citation2009), although S. buchananii has fared little better with listings ranging from ‘Insufficiently Known’, ‘Local’ and ‘Range Restricted’ to the current listing of ‘Nationally Critical’ (Given Citation1981, Citation1990; Cameron et al. Citation1993, Citation1995; de Lange et al. Citation1999, Citation2004, Citation2009).
After its formal description, S. laxa was not recorded again until it was discovered at Castle Rock, Old Man Range, Otago, in 1969. More than two decades later, in June 1991, BPJ Molloy chanced upon a small roadside population of this species growing under a limestone overhang near Ngapara, North Otago. These discoveries prompted a more thorough assessment of the species in the wild, which confirmed that it was still present at Castle Rock and Ngapara, and that it also occurred very locally in scattered sites near Macraes and Nenthorn (Johnson Citation1995). Since then, further populations have been discovered in Otago including a further 20 populations in the Macraes and Nenthorn areas, at 3 O'Clock Stream and Canadian Flat along the Taieri River, and at Rough Ridge ().
Figure 1 Map of New Zealand showing sites sampled, major cities and other locations discussed in the text. Squares, major cities; circles Simplicia laxa; triangles, Simplicia buchananii.
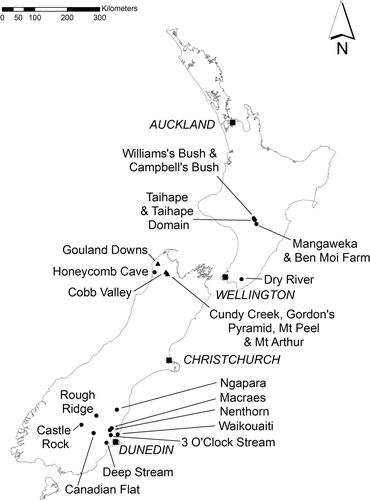
In 2005, one of us (CCO) discovered Simplicia at four sites in alluvial forest remnants near Mangaweka and Taihape in the North Island (Ogle Citation2010). These gatherings have some features suggestive of S. buchananii, a seemingly glabrous ligule, leaf-sheath and leaves, and a scabrid lemma. However, their inflorescences vary from a contracted more or less linear arrangement to a loosely spreading pyramidal shape. In this respect they match S. laxa.
Therefore, as opinions varied as to which of the two species these North Island plants should be referred (Ogle Citation2010), and because the genus as a whole is now considered threatened (see de Lange et al. Citation2009 de Lange et al. Citation2010) we undertook a reinvestigation of the genus using nuclear internal transcribed spacer (ITS), external transcribed spacer (ETS) and plastid trnL intron and trnL–trnF intergenic spacer sequence data and DNA fingerprint profiles for Simplicia using the amplified fragment length polymorphism (AFLP) technique (Vos et al. Citation1995). This had the primary aim of assessing relationships between regional groups of populations, in particular, the relationship of the newly discovered Taihape area populations to the named species S. laxa and S. buchananii, as well as the secondary aim of assessing levels of genetic variation between and within regional population groups that may be of value for conservation managers.
Materials and methods
Vouchers collected at the time DNA samples were sampled in the field, or from cultivated plants grown from these collections, are available for most locations (). No voucher was collected from the Cobb Valley S. buchananii site because plants were not in flower or fruit. However, this gathering came from a site where the species was previously gathered in 1965 (CHR 119546). The Rough Ridge sample of S. laxa was collected by Craig Wilson (Department of Conservation) in April 2009 but no voucher was preserved. AK 288071 was collected (as S. buchananii) from the Honeycomb Cave site in 2004, but an additional voucher (CHR 607318) was made from cultivated material grown at Landcare Research, Lincoln.
Table 1 Sample location, voucher and GenBank accession details for Simplicia plants used in this study.
DNA was extracted from fresh tissue using the CTAB method (Doyle & Dickson Citation1987), as described in Smissen & Heenan (Citation2007). Digestion, ligation and polymerase chain recation (PCR) amplification of DNA for AFLP fingerprinting (Vos et al. Citation1995) was conducted as described in Smissen & Heenan (Citation2007). Fragments were separated by electrophoresis on 6% polyacrylamide gels and visualized by silver staining. Because a relatively large number of samples was available for the Macraes (18), Castle Rock (20), Nenthorn (15) and Taihape (19) locations, we undertook a screen of each population group, using the primer pairs Eco–AGG with Mse–CAG and Eco–ACG with Mse–CTC, to identify duplicated genotypes and reduce the number of samples included in a final global analysis to 23 representative samples (a convenient number for our electrophoresis equipment: ). Limiting the number of samples facilitates scoring and maximizes the reproducibility of scoring because it avoids the need to compare profiles across different gels. We scored fragments in the size range of ∼50–400 bp. AFLP profiles were scored by eye to produce a binary data matrix from which simple genetic distances among individuals were calculated and subjected to UPGMA analysis using PAUP*4.0b10 (Swofford Citation2003). Bootstrap percentages were calculated from 1000 replicates.
Table 2 Samples included in AFLP analysis.
Twelve samples were selected for inclusion in a DNA sequence analysis. These represented all of the sites sampled in the study except the Macraes site. The Castle Rock site was represented in the sequencing analysis by two samples, while the other sites were represented by a single sample.
The ITS regions of the 18S–26S nrDNA spacer were amplified by PCR with the primers ITS4 and ITS5 (White et al. Citation1990) using an annealing temperature of 48°C. The plastid DNA region including the trnL intron and the trnL–trnF intergenic spacer was amplified using the primers c and f of Taberlet et al. (Citation1991) at an annealing termperature of 60°C. The ETS region of the 18S–26S nrDNA was amplified using the primers 18S-IGS (Baldwin & Markos Citation1998) and RETS4 (TGGCTACGCGAGCGCATGAG: R. Soreng personal communication) and an annealing temperature of 60°C. Sequences were deposited in GenBank (see accession numbers in ). Thirty cycles of PCR were carried out after an initial denaturation step of 120 s at 95°C. Denaturing steps were 30 s at 95°C, annealing steps were 30 s long, and extension steps were 120 s at 72°C. Cycling was followed by a 10-min incubation at 72°C.
Outgroup sequences were selected after analysis with other Poideae sequences by R. Soreng (unpublished data). Outgroup sequences were as follows (GenBank accession in parentheses): trnL intron and trnL–F spacer: Nicoraepoa andina (DQ353971), Cinna arundinacea (EU792436) and Cinna latifolia (GQ324396); ITS: Catabrosa aquatica (EU792334), Nicoraepoa andina (EU792354), Cinna arundinacea (EU792343) and Cinna latifolia (GQ324473); ETS, Catabrosa aquatica (GQ324258), Nicoraepoa andina (GQ324275), Cinna arundinacea (GQ324260) and Cinna latifolia (GQ324261). Catabrosa aquatica was not included as an outgroup in the plastid sequence dataset because it was much less similar to the other sequences than they were to each other and did not provide an unambiguous root.
Alignment was trivial and conducted manually. Gaps were coded as binary characters using the method of Simmons & Ochoterena (Citation2000) as implemented in Fastgap1.2 (Borchsenius Citation2009). An incongruence length difference (ILD) test was conducted using PAUP*4.0b10 to assess congruence between ITS and ETS sequences, using 100 heuristic search replicates, and these sequences were subsequently combined for analysis. Plastid and nuclear sequences were analysed separately. Sequences were subjected to parsimony analysis using PAUP*4.0b10 with Branch and Bound searches (Hendy & Penny Citation1982). Most parsimonious tree sets were condensed according to the criterion minimum branch length=0. Bootstrap values were calculated from 1000 replicates using Branch and Bound searches.
Results
AFLP
Preliminary experiments with the Simplicia sample collected from Honeycomb Cave (Nelson), the sample Simplicia from Campbell's Bush (Taihape) and with three samples of S. laxa from Otago demonstrated that duplicate DNA extractions and AFLP reactions produced identical DNA fingerprint profiles for each sample. The ongoing reproducibility of AFLP reactions is further supported circumstantially by the detection of identical profiles from multiple samples collected from the same locality in several cases.
Initially, AFLP profiles for plant samples from the Taihape area (Campbell's Bush, Williams's Bush, Taihape Reserve and Ben Moi Farm populations), and the Otago Castle Rock, Nenthorn and Macraes sites, were compared to assess genetic diversity within each population and to select representative genotypes for inclusion in the final sample set. The number of samples collected from each site, the number of distinct genotypes observed and the number of samples included in the final analysis are indicated in . Although we did not include all distinct genotypes for every population, those not included differed only slightly and grouped strongly with included samples from the same site in our preliminary analysis. Therefore, their inclusion would not affect groupings among populations recovered in the final analysis.
For the reduced final set of 23 samples, 111 fragments were scored by eye from silver-stained gels, using two AFLP primer combinations. In some parts of the gels, the presence of numerous fragments of similar, but slightly different, mobility made scoring difficult; these regions of the gel were not scored. For the most part, scoring was not difficult. Of the 111 fragments scored, 97 were variable, and of these 11 were restricted to a single profile.
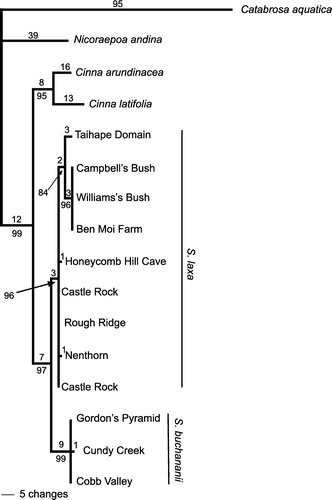
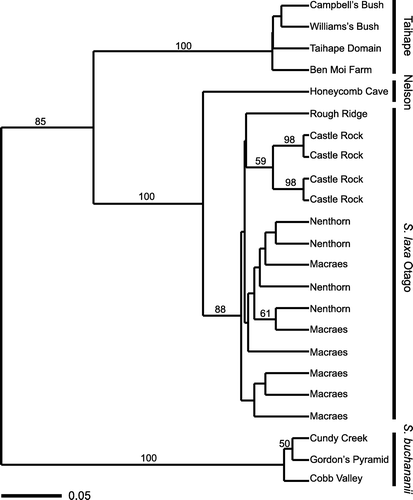
A UPGMA analysis conducted with PAUP*4.0b10 recovered clusters corresponding to the Taihape area samples, the Mt Arthur area samples and a group combining the sole Honeycomb Cave sample with all the Otago samples (). Within this last group, samples from Otago formed a subgroup. Within the Otago subgroup of samples, those from Castle Rock clustered together within a wider cluster also including the Rough Ridge, Nenthorn and Macraes samples.
Figure 2 UPGMA dendrogram for Simplicia AFLP profiles for individual plants. Bootstrap values are shown above branches where these were ≥50%.
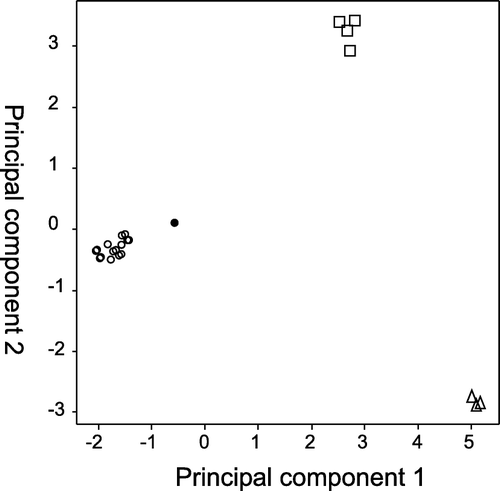
The first and second components from a principal component analysis accounted for 47 and 21% of the variation in the dataset, respectively. Subsequent components each explained ≤ 6% of the variation. The scatter plot of scores for the first two components () shows three groups corresponding with the Mt Arthur area samples, the Taihape Area samples and the Otago and Honeycomb Cave samples.
Nuclear ribosomal DNA spacer sequences (ITS and ETS)
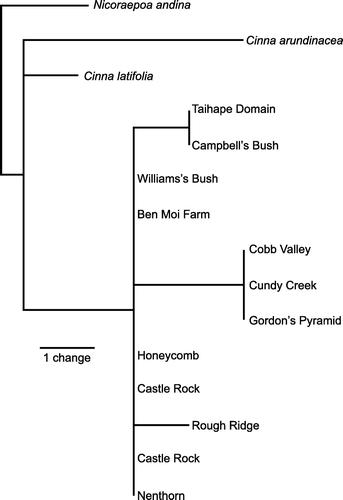
Alignment of nrDNA sequences was not problematic, requiring 7 insertions/deletions (indels) in ITS and 11 in ETS. A partition homogeneity test conducted with PAUP*4.0b10 did not find statistically significant incongruence (p=0.09) between the two regions. Inspection of the data suggests that what incongruence is present arises in the relationships among Simplicia and the outgroups, and not over ingroup relationships or placement of the root of the Simplicia tree, and is probably the result of the high divergence between the Catabrosa sequences and the other sequences. Both ITS and ETS sequences displayed numerous sites at which chromatograms indicated the presence of more than one DNA base (variation within individual plants). Parsimony analysis of the combined nrDNA sequences () provided support for clades consisting of S. buchananii samples from the Mt Arthur area (bootstrap support 99%) and S. laxa (including the Honeycomb Cave sample: bootstrap support 96%). Within S. laxa, the Taihape area samples appeared as a clade.
Plastid DNA sequences (trnL intron, trnL–trnF intergenic spacer)
There are only a few variable sites within Simplicia in this DNA region, and they are also very similar to the outgroup sequences used. After condensing branches with a minimum length of zero, a single most parsimonious tree of length 13 steps was found. Excluding uninformative characters, the most parsimonious tree had a consistency index of 0.833, retention index of 0.90 and rescaled consistency index of 0.75. Consistent with the small number of variable characters, there is little resolution in the tree (). However, a distinct chloroplast lineage was recovered from the three Mt Arthur area samples, and another from two of the four Taihape area samples.
Discussion
As is evident in and 3, the bulk of the variation in AFLP profiles is among major population groups (Taihape area, Otago, Honeycomb Cave, Mt Arthur area) and little is between or within individual localities within these. The low level of variation within each population and the identical AFLP profiles displayed by many of the plants from the same site indicate either a level of clonality and/or habitual selfing, but the dominant inheritance of AFLP markers means we cannot distinguish between these possibilities. Cultivated specimens of Simplicia set abundant seed in isolation, and the genus is regarded as self-compatible (Connor Citation1988; Edgar & Connor Citation2000).
The clustering of the four main groups of AFLP profiles {[(Honeycomb Cave, Otago area), Taihape area], Mt Arthur area}, does not reflect their geographic relationships {[(Mt Arthur area, Honeycomb Cave), Taihape area], Otago area}. It is difficult to relate the phylogeographic pattern uncovered by AFLP profiles directly to the biological species concept because the major genetic groupings are allopatric and the sampled distribution is too sparse to make statistically valid comparisons between the populations’ genetic and geographic distances.
However, accommodating the newly found Taihape area populations of Simplicia within the existing taxonomy is probably best achieved by including them within S. laxa rather than in S. buchananii, given their greater similarity in AFLP profiles to the former. Parsimony analysis of nrDNA sequences provides support (bootstrap 96%) for the monophyly of S. laxa thus circumscribed (). Sequences of ITS for several specimens of S. laxa from Macraes are identical to those from Nenthorn (J Keeling personal communication; RD Smissen unpublished data) but they are not included in the analysis presented here because ETS sequences are not available. Although the Simplicia at Honeycomb Cave has been considered as belonging to S. buchananii (SP Courtney & P Wardle in litt.; de Lange & Murray Citation2002), including by us at the outset of this study, all four genetic datasets are more consistent with it being part of S. laxa. In particular, the AFLP data provide strong support for this re-determination. Wild-gathered examples of Honeycomb Cave plants had been placed with S. buchananii because of their linear panicles, which are rather like those of S. buchananii. However, in the few wild-gathered specimens that we have seen from Honeycomb Cave (e.g. AK 252968) there is at least one of the basal panicle branches that is strongly reflexed, which is typical of S. laxa and not S. buchananii. Further, cultivated plants from Honeycomb Cave soon developed the typically lax pyramidal shape present in the mature panicle of S. laxa (see AK 288071). Despite this, Honeycomb plants are unusual within S. laxa samples that we have seen in that the adaxial leaf surface is not especially hairy, and in some specimens the lemma (especially those that are over-mature) is covered in prickle-teeth rather than the hairs typical of the majority of Otago populations of S. laxa (e.g. AK 304845, AK 304847). In these respects, Honeycomb Cave plants approach North Island specimens of S. laxa.
On the basis of these discoveries it is clear that S. laxa is both genetically and morphologically variable. AFLP profiles readily divide the species into two major genetic groups, one comprising the populations near Taihape, and the other the South Island populations (Honeycomb Cave and Otago). Theoretically, our genetic data are quite consistent with recognizing each of these genetic groups as either species or subspecies. However, finding a clear morphological basis on which to further separate the S. laxa genetic groupings has proved difficult.
Pending a detailed and thorough characterization of morphological variation in S. laxa, we offer the following observations based primarily on examination of herbarium specimens at WELT, CHR and AK by PJ de Lange. The majority of Otago S. laxa possess copious, long, patent sericeous hairs on the leaf sheaths and adaxial leaf surface, conspicuous hairy ligules and hairy lemma. This departs from the sole condition seen in Taihape populations in which the leaf sheaths and leaves are sparsely and minutely hairy tending toward glabrate or even glabrous (though the lower leaf sheaths, near the soil surface, are densely covered in minute retrorse hairs in glabrous plants; see AK 304804, AK 304805), a glabrous ligule and the lemma sparsely to copiously covered in short prickle-teeth. These distinctions potentially serve as a useful basis for taxonomic segregation of S. laxa into northern and southern groups, except that plants at Honeycomb Cave display intergrading features (e.g. AK 252968, AK 288071) and plants from Ngapara in North Otago (65 km north of Waikouaiti) are an exact match for Taihape plants (e.g. AK 208577, AK 288070). Moreover, although plants from Ngapara were unavailable for our AFLP study, an unpublished ITS sequence (J Keeling personal communication) from a plant collected there (AK 285424) clusters with those from Taihape (not shown) suggesting that the genetic groups may not be as well separated geographically as our AFLP analysis implies. A further problem is that the type suite of S. laxa shows an apparent continuum between typical ‘hairy’ Otago and ‘glabrate’ Taihape plants.
The type material was gathered from Waikouaiti on the north-eastern coastline of Otago, where the species is possibly now extinct (Johnson Citation1995). The gatherings from here are copious and it is possible that more than one individual is involved. For example, the lectotype from Waikouaiti (WELT SP043017) is glabrate and in many respects a close match for the'glabrate’ Taihape plants, whereas the isolectotype (WELT SP043021) is extremely hairy like the majority of Otago plants. Because these patterns of variation are complex they require further study before definite conclusions about the taxonomic status of ‘glabrate’ versus ‘hairy’ S. laxa populations can be made. Therefore, while acknowledging the genetic partitioning of S. laxa evident from this study, we feel that, at this stage in our research, it is best to retain the two species of Simplicia, S. buchananii and S. laxa, while accepting that S. laxa is morphologically variable.
Taxonomic conclusions
Current treatment of Simplicia as two species, S. buchananii and S. laxa, is consistent with morphological and genetic evidence. However, AFLP and DNA sequence data strongly suggest that the Simplicia at Honeycomb Cave (Nelson) is S. laxa, not S. buchananii. This result is also consistent with reappraisal of the morphological characters. North Island plants from the Taihape area should also be excluded from S. buchananii and are better treated as part of S. laxa. Based on their morphology, historical gatherings from the south-eastern Wairarapa of the North Island are also best placed within S. laxa. Rediscovery of Simplicia in the Wairarapa would allow additional genetic work to help clarify the taxonomic significance of genetic variation within S. laxa. Further AFLP analysis including plants from Ngapara could also be undertaken to test the relationship with Taihape plants suggested by their morphology and ITS sequences.
Conservation
From a conservation perspective, S. buchananii remains something of an enigma. The species is treated as ‘Threatened/Nationally Critical’ qualified DP (Data Poor), RR (Range Restricted), Sp (Sparse) by de Lange et al. (Citation2009), and this threat status is probably accurate. Although it is clear that S. buchananii is a naturally uncommon biologically sparse grass of montane karst habitats, and that all of the known populations are secure within Kahurangi National Park, during the sampling for this study it was recognized that populations are often very small (in the tens rather than hundreds of plants). Further, it was observed that at some S. buchananii sites aggressive weeds such as Holcus lanatus L. and Mycelis muralis (L.) Dumort are now present and these species are likely to displace Simplicia unless they are actively controlled. For these reasons, we see no need to change the current threat status of this species.
Although this study has shown that S. laxa is much more widespread than its montane sister species S. buchananii, the current threat status of ‘Threatened/Nationally Critical’ qualified CD (Conservation Dependent), Sp (Sparse) is probably still appropriate. None of the North Island populations is secure from weed invasion and most occur in small, seriously degraded, privately owned, forest remnants. Nevertheless, with the recognition that S. laxa is really a species of forested habitats (Ogle Citation2010), rather than being ‘a species of relatively sheltered, mainly east- or south-aspect rock outcrops where the specific micro-site is the immediate base of the rock outcrop or the floor of small caves’ (see Johnson Citation1995), we are optimistic that further S. laxa populations will be found following diligent searching within the types of habitats described by Ogle (Citation2010).
Acknowledgements
We thank Phil Knightbridge, Rhys Gardner, Henry Connor, Peter Johnson, Jeanette Keeling, Vivienne McGlynn, Graeme La Cock, Phyllis Leigh, John Barkla, Craig Wilson, Peter Heenan, Gary Houliston, Christine Bezar, Lynn Gillespie and Robert Soreng for providing specimens, unpublished data, field assistance, advice or comments on the manuscript. We thank David Purcell (Landcare Research) and Geoff Davidson (Oratia Native Plant Nurseries) for maintaining the live Simplicia collections used for this study. We thank Fran Thorsen for designing and drafting . This work was supported by funding from the Department of Conservation, Research and Development Conservation Management Units Fund and the New Zealand Foundation for Research, Science and Technology through backbone funding of the ‘Defining New Zealand's Land Biota’ programme. This paper also benefited from constructive input by two anonymous reviewers.
References
- Baldwin , BG and Markos , S . 1998 . Phylogenetic utility of the external transcribed spacer (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae) . Molecular Phylogenetics and Evolution , 10 : 449 – 463 .
- Borchsenius F 2009 . FastGap 1.2 . Department of Biological Sciences, University of Aarhus, Denmark. Published online at http://192.38.46.42/aubot/fb/FastGap_home.htm (accessed 29 July 2010 ).
- Buchanan J 1880 . The indigenous grasses of New Zealand . Wellington, , New Zealand , James Hughes .
- Cameron , EK , de Lange , PJ , Given , DR , Johnson , PN and Ogle , CC . 1993 . New Zealand Botanical Society threatened and local plant lists (1993 revision) . New Zealand Botanical Society Newsletter , 32 : 14 – 28 .
- Cameron , EK , de Lange , PJ , Given , DR , Johnson , PN and Ogle , CC . 1995 . New Zealand Botanical Society threatened and local plant lists (1995 revision) . New Zealand Botanical Society Newsletter , 32 : 15 – 28 .
- Connor , HE . 1988 . Breeding systems in New Zealand grasses X. Species at risk for conservation . New Zealand Journal of Botany , 26 : 163 – 167 .
- de Lange , PJ , Heenan , PB , Given , DR , Norton , DA , Ogle , CC , Johnson , PN and Cameron , EK . 1999 . Threatened and uncommon plants of New Zealand . New Zealand Journal of Botany , 37 : 603 – 628 .
- de Lange PJ , Heenan PB , Norton DA , Rolfe JR , Sawyer JWD 2010 . Threatened plants of New Zealand . Christchurch, , New Zealand , Canterbury University Press .
- de Lange , PJ and Murray , BG . 2002 . Contributions to a chromosome atlas of the New Zealand flora – 37. Miscellaneous families . New Zealand Journal of Botany , 40 : 1 – 23 .
- de Lange , PJ , Norton , DA , Courtney , SP , Heenan , PB , Barkla , JW , Cameron , EK , Hitchmough , R and Townsend , AJ . 2009 . Threatened and uncommon plants of New Zealand (1998 revision) . New Zealand Journal of Botany , 47 : 61 – 96 .
- de Lange , PJ , Norton , DA , Heenan , PB , Courtney , SP , Molloy , BPJ , Ogle , CC , Rance , BD , Johnson , PN and Hitchmough , R . 2004 . Threatened and uncommon plants of New Zealand . New Zealand Journal of Botany , 42 : 45 – 76 .
- Doyle , JJ and Dickson , EE . 1987 . Preservation of plant samples for DNA restriction endonuclease analysis . Taxon , 36 : 715 – 722 .
- Edgar E , Connor HE 2000 . Flora of New Zealand volume V: Gramineae . Lincoln, , New Zealand , Manaaki Whenua Press .
- Given , DR . 1976 . A register of rare and endangered indigenous plants in New Zealand . New Zealand Journal of Botany , 14 : 135 – 149 .
- Given DR 1981 . Rare and endangered plants of New Zealand . Wellington, , New Zealand , AH & AW Reed .
- Given DR 1990 . Threatened and local plant lists – New Zealand Botanical Region . Christchurch, , New Zealand , Botany Institute, DSIR Land Resources .
- Hendy , MD and Penny , D. 1982 . Branch and bound algorithms to determine minimal evolutionary trees . Mathematical Biosciences , 59 : 277 – 290 .
- Johnson PN 1995 . The rare grass Simplicia laxa – field status, ecology and conservation. Science for Conservation 15 . Wellington, , New Zealand , Department of Conservation .
- Kirk , T . 1897 . Description of a new genus of Gramineae . Transactions and Proceedings of the New Zealand Institute , 29 : 497
- Ogle , CC . 2010 . Rediscovery of a rare species of grass in the genus Simplicia, in the North Island . Wellington Botanical Society Bulletin , 51 : 38 – 46 .
- Simmons , MO and Ochoterena , H . 2000 . Gaps as characters in sequence-based phylogenetic analysis . Systematic Biology , 49 : 369 – 381 .
- Smissen , RD and Heenan , PB . 2007 . DNA fingerprinting supports hybridisation as a factor explaining complex natural variation in Phormium (Hemerocallidaceae) . New Zealand Journal of Botany , 45 : 419 – 432 .
- Swofford DL 2003 . PAUP* . Phylogenetic Analysis Using Parsimony (*and other methods). Version 4 . Sunderland, MA , Sinauer Associates .
- Taberlet , P , Gielly , G , Pautou , G and Bouvet , J . 1991 . Universal primers for the amplification of three non-coding regions of chloroplast DNA . Plant Molecular Biology , 17 : 1105 – 1109 .
- Vos , P , Hogers , R , Bleeker , M , Reijans , M , van de Lee , T , Hornes , M , Frijters , A , Pot , J , Peleman , J , Kuiper , M and Zabeau , M . 1995 . AFLP: a new technique for DNA fingerprinting . Nucleic Acids Research , 23 : 4407 – 4014 .
- White TJ , Bruns T , Lee S , Taylor J 1990 . Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics . In: Innis M , Gelfand D , Sninsky J , White T PCR protocols: a guide to methods and applications . San Diego, CA , Academic Press . Pp. 315 322 .
- Williams GR , Given DR 1981 . The red data book of New Zealand . Wellington, , New Zealand , Nature Conservation Council .
- Wilson CM , Given DR 1989 . Threatened plants of New Zealand. DSIR field guide . Wellington, , New Zealand , DSIR Publishing .
- Zotov , VD . 1943 . Certain changes in the nomenclature of New Zealand species of Gramineae . Transactions and Proceedings of the Royal Society of New Zealand , 73 : 233 – 238 .
- Zotov , VD . 1971 . Simplicia T. Kirk (Gramineae) . New Zealand Journal of Botany , 9 : 539 – 544 .