Abstract
Twelve polymorphic simple sequence repeat markers were developed from transcribed sequences generated by RNA-seq. These were then used to assess relationships among selected putative species of Craspedia in Kahurangi National Park, New Zealand, including three putative species in sympatry at Mt Arthur; plants from two putative species from the nearby Mt Mytton; and plants from the Marino Mountains, where at least four putative species have been listed. We confirmed that two of the putative species present on Mt Arthur are also present on Mt Mytton, but identified another Craspedia there that was previously not recognised as being distinct. At the Marino Mountains the situation appears more complex. One putative species there is clearly distinct from all other plants sampled. However, the remainder of the Marino Mountains plants could not be assigned to genetic groups consistent with the putative species, nor could we clearly relate them to the putative species from Mt Arthur or Mt Mytton.
Introduction
The species of Craspedia G. Forst. (Asteraceae, tribe Gnaphalieae, subtribe Angianthinae) are conspicuous members of many plant communities in Australia and New Zealand. Like other species of subtribe Angianthinae they are characterised by an unusual double composite inflorescence. Craspedia is the only genus of the predominantly Australian Angianthinae that occurs in New Zealand. Its New Zealand species constitute a monophyletic group (Ford et al. Citation2007). On the basis of phylogenies from chloroplast and nuclear DNA sequences, Ford et al. (Citation2007) concluded that New Zealand species are derived from within an Australian radiation of Craspedia. The low levels of sequence divergence among species of Craspedia in New Zealand reported by Ford et al. (Citation2007) suggest they radiated recently.
Allan (Citation1961) recorded six species of Craspedia in New Zealand from a small number of available herbarium specimens. However, Druce (Citation1993), who made many field studies and collections, suggested much greater diversity and listed more than 45 informally named putative species that might or might not warrant taxonomic recognition. Breitwieser et al. (Citation2010) explained that much difficulty in the delimitation of species is caused by characters that lack clear discontinuity in variation, are predominantly quantitative rather than qualitative, and are environmentally influenced. Although Craspedia has numerous ill-defined forms, those that grow at the same site can often be distinguished easily from one another. Glasshouse observations (I Breitwieser & KA Ford unpubl. data) and available genetic evidence (Breitwieser et al. Citation2010 and unpubl. data) suggest that outcrossing is the norm for most Craspedia and available chromosome counts suggest a uniform 2n = 22 (Dawson Citation1995; Dawson et al. Citation1999). Therefore, our research seeks to test whether broadly sympatric putative species (i.e. those with overlapping ranges), and especially those that grow in syntopy (i.e. at the same site), are reproductively isolated from each other. We consider reproductive isolation in sympatry to be necessary (but not always sufficient) to recognise putative species as distinct taxonomic species.
A previous study of genetic variation among and within three putative species of Craspedia that grow together at the same site at Mt Arthur (Kahurangi National Park, Nelson Province) and are distinguishable by their morphology was conducted by Breitwieser et al. (Citation2010) using amplified fragment length polymorphism (AFLP) (Vos et al. Citation1995). That study recovered three genetic clusters consistent with morphologically defined groups (Craspedia sp. 1 with informal name ‘Craspedia elongata’, CHR 514388; Craspedia sp. 2 with informal name ‘Craspedia long hairs’, CHR 514387; Craspedia sp. 3 with informal name ‘Craspedia marble’, CHR 514389). No admixture among these genetic clusters was detected. However, we have found that AFLP profiles perform poorly when applied to more diverse sets of samples (e.g. wider samplings of putative species representing greater genetic diversity) largely as a result of homoplasy, but also problems with reproducibility over months or years. Therefore, we set out to characterise simple sequence repeat (SSR or microsatellite) markers for use in New Zealand Craspedia. These are often more reproducible than AFLPs, are co-dominant (AFLP markers are generally used as dominant markers), and allow greater flexibility in terms of sampling and data collection (Nybom et al. Citation2014).
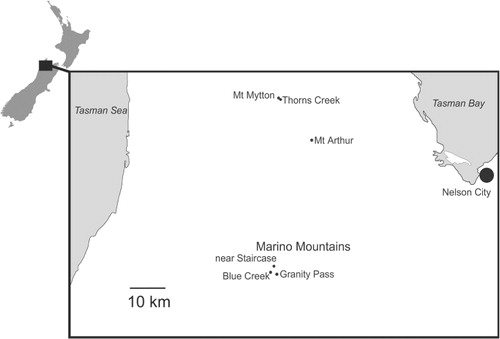
Here, we report the characterisation of 12 SSR markers that are widely applicable to Craspedia from New Zealand (I Breitwieser et al., unpubl.). We also test these markers by examining the same three putative species previously examined at Mt Arthur using AFLP and expand sampling to plants from Mt Mytton and the Marino Mountains (all in Kahurangi National Park, Nelson Province: ). The majority of the putative species of Craspedia (Druce Citation1993) are from Kahurangi National Park. This area is unique in New Zealand for its complex geological features and is part of an area of high species-level plant diversity (Rogers & Walker Citation2005). Uplifted limestone and marble deposits form a discontinuous belt up to 1200 m thick extending from Takaka to Mt Arthur to the Marino Mountains (Lee & Nelson Citation2008). In particular, these limestone and marble ecosystems are botanically very rich. Although we generally try to avoid using informal names, in order to avoid confusion in this paper, we are enclosing such names in double quotes, as recommended in the guidelines for authors.
Table 1 Localities and putative informally named Craspedia species.
The three morphologically distinct putative species present at Mt Arthur are referred to in Breitwieser et al. (Citation2010) as Craspedia sp. 1, C. sp. 2 and C. sp. 3, but are here referred to by informal names ‘C. elongata’, ‘C. long hairs’ and ‘C. marble’, respectively (). Craspedia sp. 1 was described by Allan (Citation1961) as Craspedia lanata var. elongata Allan and listed by Druce (Citation1993) informally at species rank as ‘C. elongata’, thus suggesting that its rank needs further study. However, our most recent results from SSR analyses indicate that the plants of C. sp. 1 on Mt Arthur might not belong to the same taxon as plants from Lake Tennyson, Canterbury Province, the type locality of C. lanata var. elongata (I Breitwieser et al., unpubl.). Craspedia sp. 2 is listed by Druce (Citation1993) as ‘C. long hairs’ and C. sp. 3 as ‘C. calcicole’, although he also annotated herbarium sheets from this site with the informal name ‘C. marble’. Druce did not provide any characters that distinguish ‘C. marble’ from ‘C. calcicole’, but our studies indicate that the difference seems to be mainly in the leaf shape and a character of the cypsela. The length-to-width ratio of the rosette leaves of ‘C. calcicole’ is usually larger than in ‘C. marble’, and, whereas the cypselae of ‘C. calcicole’ have infinite bifid hairs, those of ‘C. marble’ are few and very short and the cypsela surfaces are densely covered in glandular twin hairs and copious mucilage (Breitwieser et al., unpubl.). The plants of C. sp. 3 at Mt Arthur have the characteristics of ‘C. marble’, and this is the informal name we use here. However, the most obvious distinguishing characters of the three putative species of Craspedia at Mt Arthur are based on the appearance of their rosette leaves. ‘Craspedia elongata’ can be distinguished by the dense, more or less appressed, greyish white tomentum of cottony hairs, ‘C. long hairs’ by the hispid non-glandular, multicellular hairs up to 2 mm long and faint white leaf margin, and ‘C. marble’ by stalked glandular hairs up to 0.3 mm long and distinctly white leaf margins of tangled cottony hairs ().
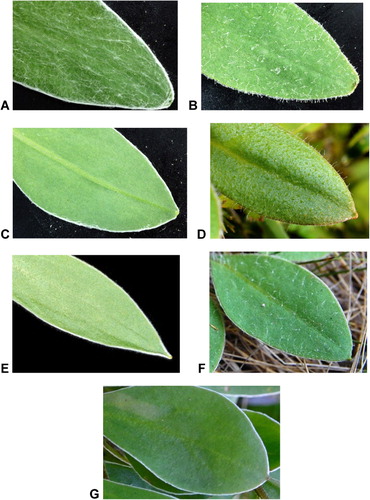
On Mt Mytton we collected two putative species from the limestone area of that locality. One of these putative species was given the informal name ‘Craspedia Mytton’ by Druce (Citation1993), whereas the other corresponds to ‘C. long hairs’ from Mt Arthur. ‘Craspedia Mytton’ is morphologically very similar to ‘C. marble’ from Mt Arthur. We have not found any information in Druce's notes, lists or herbarium sheets on why he gave it a different informal name, nor have our own studies identified significant morphological differences. We also collected plants of Craspedia (CHR 635119) in the basin of Thorns Creek, just below the limestone area of Mt Mytton. They are very similar to the ‘C. long hairs’ we collected in the limestone area of Mt Mytton. On the basis of herbarium information and morphological characters, we identified them as ‘C. long hairs’.
On the Marino Mountains, using Druce's list of informally named putative species of Craspedia (Druce Citation1993) as our guide, we sampled four morphologically different putative species (see ). One of these (CHR 635113) corresponds to ‘C. elongata’ from Mt Arthur and one (CHR 635117) to ‘C. long hairs’. These are the plants with dense, more or less appressed, greyish white tomentum of cottony hairs on the rosette leaves (‘C. elongata’) and the plants with hispid non-glandular, multicellular hairs up to 2 mm long on the rosette leaves and faint white leaf margin (‘C. long hairs’), respectively (). The third form (CHR 635120) is similar to ‘C. marble’ from Mt Arthur in having stalked glandular hairs up to 0.3 mm long on the rosette leaves and distinctly white leaf margins of tangled cottony hairs, but can be referred to ‘C. calcicole’ because of its leaf ratio and fruit characteristics. The fourth form on the Marino Mountains is listed by Druce (Citation1993) as ‘C. Owen’ (CHR 635116), with the informal name referring to Mt Owen, the highest peak in the Marino Mountains and of Kahurangi National Park. He did not provide any distinguishing characters. From our studies, this form is characterised by rosette leaves with multicellular hairs with cottony tips up to 6 mm long and stalked glandular hairs up to 0.3 mm long, and white leaf margins of short multicellular hairs, c. 1 mm long, with long cottony tips. The cypselae are densely covered with glandular twin hairs and copious mucilage, and bifid hairs are absent (I Breitwieser et al., unpubl.).
Here, we seek to answer the following questions using the 12 SSR markers:
Can the three putative species ‘C. elongata’, ‘C. long hairs’ and ‘C. marble’ that we confirmed from Mt Arthur (Breitwieser et al. Citation2010) as Craspedia sp. 1, C. sp. 2 and C. sp. 3, respectively, be retrieved? This would allow us to assess the performance of the SSR markers.
Can ‘C. Mytton’ from Mt Mytton be distinguished genetically from ‘C. marble’ from Mt Arthur, and can ‘C. calcicole’ from the Marino Mountains be distinguished from these?
Can ‘C. long hairs’ from Mt Arthur, the Marino Mountains and Mt Mytton be distinguished?
Can plants of ‘C. elongata’ from the Marino Mountains be distinguished from those at Mt Arthur?
Is ‘C. Owen’ from the Marino Mountains distinct in comparison to all the other forms examined?
Materials and methods
Materials
This study tests putative species of New Zealand Craspedia from Mt Arthur, Mt Mytton and the Marino Mountains, all in Kahurangi National Park (). Material was collected by the authors in Kahurangi National Park at Mt Arthur (−41.219175°S, 172.687911°E), Mt Mytton (limestone area −41.121455°S, 172.583117°E; the basin of Thorns Creek −41.124372°S, 172.589068°E); and the Marino Mountains (near the Staircase −41.509905°S, 172.565369°E; Granity Pass −41.528316°S, 172.574955°E; Blue Creek above Granity Pass Hut −41.523823°S, 172.554189°E). Vouchers for the forms from each mountain are deposited at CHR ().
We sampled leaf material from 10 plants of each putative species at each site when they were abundant or fewer samples in small populations. We collected plants from all habitats occurring at the site and sought to capture the whole visible variation of each form. On Mt Arthur we sampled ‘C. elongata’, ‘C. long hairs’ and ‘C. marble’ at the same sites as for our AFLP studies (Breitwieser et al. Citation2010). On Mt Mytton we sampled ‘C. Mytton’ and ‘C. long hairs’ from the limestone area where the two putative species grow together. However, ‘C. long hairs’ was uncommon at that site. In the basin of Thorns Creek, lower down on Mt Mytton, ‘C. long hairs’ was very common. All plants of Craspedia in the basin of Thorns Creek are morphologically uniform. On the Marino Mountains, we collected at three nearby sites. At the site near the Staircase, ‘C. calcicole’, ‘C. elongata’ and ‘C. Owen’ grow together. At Granity Pass some plants seemed to be intermediate between plants that we identified as ‘C. elongata’ and ‘C. calcicole’, although ‘C. Owen’ was morphologically distinct and easy to identify. At the third site, Blue Creek, above Granity Pass Hut, ‘C. Owen’ is also morphologically distinct. We identified those plants with the typical cottony leaf hairs as ‘C. elongata’, but had problems distinguishing many plants of ‘C. calcicole’ from ‘C. long hairs’ because they looked so similar. We could do this only on the basis of a leaf hair character: ‘C. calcicole’ has stalked glandular hairs up to 0.3 mm long, whereas ‘C. long hairs’ has multicellular hairs up to c. 2 mm long.
SSR characterisation
Approximately 250 mg of leaf tissue was excised from a glasshouse-grown Craspedia plant (voucher CHR 554374) and mRNA extracted using an Invitrogen mRNA direct™ kit (catalogue number 610.11). A cDNA library was then produced using the cDNA Rapid Library Preparation Method – GS Junior Titanium Series (May 2010). The cDNA library was sequenced by the GS Junior system following emPCR using a GS Junior Titanium emPCR kit (Lib-L) kit (catalogue no. 05996481001). Processed sequence output from the 454 sequencer was searched for SSR motifs using Microsatellite Commander (Rozen & Skaletsky Citation2000; Faircloth Citation2008). Primers to amplify selected repeat-containing regions were designed with one of each pair incorporating a 5′-tail matching the M13 forward primer sequence (TGTAAAACGACGGCCAGT) to allow incorporation of fluorescent labels (Boutin-Ganache et al. Citation2001).
We screened 52 primer pairs against a panel of 12 Craspedia samples from a range of appearances and provenances to identify those that amplified polymorphic profiles consistent with the amplification of a single locus. These criteria were met by 12 primer pairs. Polymerase chain reaction (PCR) conditions were initial denaturation for 2 min at 95 °C, followed by 15 cycles of PCR for 30 s at 95 °C then 30 s at the primer-specific annealing temperature shown in , and 60 s at 72 °C, then a further 20 cycles with the annealing temperature lowered to 49 °C to promote incorporation of the fluorescent label. Annealing temperatures for each primer pair were selected after gradient PCR on an Eppendorf Mastercycler (Hamburg, Germany) with annealing temperatures varying from 50 to 70 °C.
Table 2 Details of the simple sequence repeat markers characterised in this study.
Genotyping
DNA for genotyping was extracted from fresh tissue using the CTAB method (Doyle & Dickson Citation1987) as described in Smissen & Heenan (Citation2007) or by magnetic bead capture using the Maxwell system (Promega).
Fragment size analysis was conducted with an ABI 3500XL machine using ROX-labelled GS500 size standard. Allele scoring was undertaken manually using GeneMarker 1.51 (SoftGenetics, State College, PA, USA) to visualise fragment size analysis files. Occasional reactions failing to produce a detectable fragment were repeated.
Analysis of genotypes
We have adopted a two-stage process for the analysis of our genotype data. This aimed first to assign individuals from a collecting site to genetic populations and then to examine relationships among those populations. Genotypes of Craspedia from Mt Arthur, Mt Mytton and the Marino Mountains were analysed separately using Structure 2.3.3 (Pritchard et al. Citation2000; Falush et al. Citation2003) to determine the number of genetic populations present by examining plots of ΔK (Evanno et al. Citation2005) and log probability scores (lnPr(X|K)) for differing values of K. Samples from Mt Arthur and Mt Mytton together and from all the sites together were also subjected to Structure analyses. For Structure analyses we selected options for a model of admixture among populations with allele frequencies correlated among them as recommended by the authors of the software. A Markov chain Monte Carlo (MCMC) method was run for 100,000 generations with the first 20,000 discarded as burn-in. The number of populations modelled (K) was varied from 1 to 15 with 10 replicate runs at each value and the results analysed with Structure Harvester (Earl & von Holdt Citation2012). Other settings were default. Summary statistics, population genetic distances among populations and neighbour-joining trees (Saitou & Nei Citation1987) were generated using Powermarker 3.25 (Liu & Muse Citation2005) using the genetic distance model of Nei (Citation1972).
Results
Primer sequences, annealing temperature, repeat sequence and the observed length-variation for the 12 markers employed in this study are shown in . Summary statistics for the variation observed are shown in . At least one fragment could be amplified for most markers from most samples, except for markers Cras15 and Cras36 (availability scores of 0.89 and 0.88, respectively; ). These low availability scores may reflect the presence of null alleles. This is particularly the case for Cras36, all but one of the samples that did not yield any fragments with this marker came from the Marino Mountains suggesting a null allele is common there but rare or absent at the other sites.
Table 3 Summary statistics for SSR markers characterised in this study.
Estimation of K, the number of populations
The values of K with the highest values of ΔK and log probability scores (lnPr(X|K)) for each set of samples analysed are shown in . Maximum numbers for both quantities were obtained for the same value of K for the analysis of Mt Arthur samples alone (K = 3) and for the analysis of all the samples (K = 6). For the Mt Mytton samples, a prominent peak in ΔK was observed at K = 3 but the highest value was at K = 12, whereas lnPr(X|K) was highest for K = 3. The high value of ΔK observed at K = 12 appears to be the result of a particularly low mean and high variance for lnPr(X|K) at K = 13 rather than a particularly good fit of the model at K = 12. Two of the runs at K = 13 gave much lower values of lnPr(X|K) than the remainder, suggesting that they had not converged. When Mt Arthur and Mt Mytton samples were analysed together, the highest value of ΔK was at K = 4 and the highest value of mean lnPr(X|K) was at K = 5. However, lnPr(X|K) at K = 5 was only marginally better than at K = 4 (−1549.62 versus −1556.00) and came at the expense of a considerable increase in the variance (14.97 versus 0.58). For the Marino Mountains samples, the highest value of ΔK was at K = 2, whereas the highest value of lnPr(X|K) was at K = 5. In this case, mean values of lnPr(X|K) are notably higher at K = 3 to K = 5 than at K = 2, and the variances increase sharply above K = 3 (). Given the potential for mild departures from the model to lead to an overestimate of K (Pritchard et al. Citation2010), we have chosen to show Structure bar-plots for the lower value of K when different values were suggested by ΔK and lnPr(X|K) (), with the exception of the Marino Mountains samples where we have shown bar-plots for both K = 2 and K = 3 (–, respectively).
Table 4 Values of K (the number of populations) for each structure analysis undertaken as indicated by comparison of ΔK and mean lnPr(X|K).
Grouping individuals into populations at each site
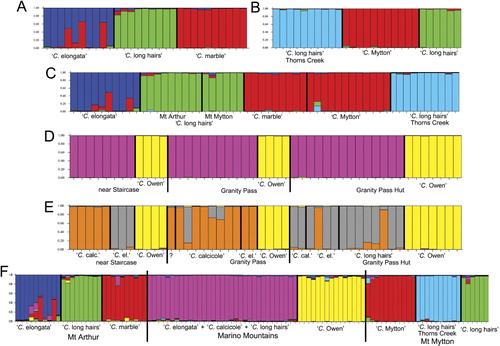
At Mt Arthur, Structure recovered three populations consistent with field identifications of the Craspedia plants (). Although, at face value, the Structure bar-plot suggests that there may be some admixture among two of the populations, 90% probability intervals (not shown) for these samples encompass pure parentage from the population corresponding with their field ID. That is, there is only weak evidence in our data to suggest that they are hybrids and a conservative interpretation is that they may be hybrids or may be pure.
Our data strongly suggest that there are three () species at Mt Mytton rather than the two we hypothesised at the outset of this study. Structure runs with K = 2 attribute samples to groups that are inconsistent with our hypothesis, as the samples from the basin of Thorns Creek (collected as ‘C. long hairs’) are separated from the remainder of the samples (collected as ‘C. Mytton’ and ‘C. long hairs’). With K = 3, one group corresponded to ‘C. Mytton’, whereas ‘C. long hairs’ from the limestone area and from the basin of Thorns Creek were recovered as two different populations.
Our results for the Marino Mountains departed considerably from our expectations. First, comparison of ΔK values from Structure analyses suggested the presence of two populations rather than the four putative species suggested by Druce (Citation1993). One of the populations suggested by Structure corresponds exactly to the plants we collected as ‘C. Owen’, whereas the other population suggested by Structure included all the remaining Marino Mountains collections (). If the value of K is increased to 3 () or more (bar-plots not shown) then some division of this group is achieved, but it is still not consistent with assignment of the samples to Druce's (Citation1993) informally named putative species except at one of the three Marino Mountains sites (near Staircase). Instead, with K = 3, Structure suggests that the samples we collected as ‘C. elongata’, ‘C. long hairs’ and ‘C. calcicole’ were members of two populations and perhaps their hybrids, with limited correlation with morphology-based field identification according to Druce (Citation1993). With K = 4 or K = 5 the grouping of ‘C. Owen’ is maintained, whereas the remaining samples show varying proportions of ancestry from each of the other four hypothetical populations.
Grouping individuals into populations across sites
Structure analysis of genotypes from Mt Arthur and Mt Mytton together support Druce's hypothesis (1993 and herbarium sheet annotations) that ‘C. long hairs’ collected on Mt Arthur also occurs on the limestone area of Mt Mytton, but does not support the hypothesis that ‘C. marble’ from Mt Arthur and ‘C. Mytton’ from Mt Mytton are two species (). Our data cannot distinguish them. However, the plants we collected as ‘C. long hairs’ from the basin of Thorns Creek at Mt Mytton are suggested by Structure to be unrelated to ‘C. long hairs’ from the limestone area on Mt Mytton and from Mt Arthur (). This was an entirely unexpected result.
When samples from all three areas are analysed together using Structure the results maintain the groupings observed in the more restricted analyses for each area and do not associate any of the plants from the Marino Mountains with any of those from Mt Arthur or Mt Mytton (K = 6; ).
Relationships among populations
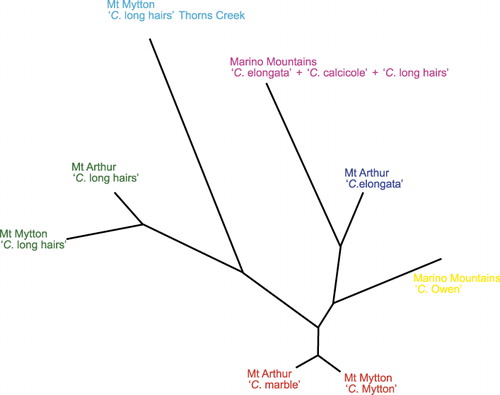
Individual genotypes were grouped into populations on the basis of their site of origin and the results of the Structure analyses and relationships among these populations explored with a neighbour-joining analysis using the genetic distance model of Nei (Citation1972). The neighbour-joining tree () illustrates the close relationship between the Mt Arthur and Mt Mytton populations of ‘Craspedia long hairs’, as well as populations of ‘C. marble’ and ‘C. Mytton’ from these sites. Conversely, none of the Marino Mountain populations seem closely related to any of the populations at Mt Arthur or Mt Mytton. However, in our neighbour-joining tree, one of the populations from the Marino Mountains is most similar to ‘C. elongata’ from Mt Arthur.
Discussion
Our analyses are aimed at testing whether the putative species of Craspedia we sampled constitute distinct genetic populations. This is a minimal test of whether they should be considered taxonomic species. That is, populations found by our Structure analyses are not necessarily distinct species but might constitute distinct populations within a species. The results we report here are consistent with those previously reported for an AFLP study of Mt Arthur Craspedia and confirm the presence of three reproductively isolated populations of Craspedia there. In contrast to the AFLP study, our SSR data suggest some hybridisation between two of the three forms (‘C. marble’ and ‘C. elongata’; ) might occur there. However, 90% confidence intervals for ancestry estimates include ‘pure’ ‘C. elongata’ or ‘C. marble’ for all of these samples. Moreover, when Mt Arthur samples are analysed with Mt Mytton samples () some improvement in separation of ‘C. elongata’ and ‘C. marble’ is achieved. Therefore, this is at best weak evidence for the presence of hybrids at Mt Arthur.
Our results also confirmed our expectations that ‘C. long hairs’ is present on Mt Arthur and Mt Mytton and that ‘C. marble’ on Mt Arthur and ‘C. Mytton’ on Mt Mytton belong to the same putative species, but upset our expectations by suggesting the plants from Thorns Creek we had considered as part of ‘C. long hairs’ are members of a population that is genetically distinct from the other putative species we examined (voucher of this population: CHR 635119). This result is significant, because it suggests that the selection of morphological characters, with emphasis on rosette leaf characters that Druce et al. (Citation1987) used to distinguish many of the putative species in Craspedia, might not be able to distinguish some biological species.
The greatest discrepancy in identifications based on Druce (Citation1993, herbarium sheet annotations) and our genetic data relates to the Marino Mountains collections. One of the putative species identified a priori was genetically well differentiated from all other collections from all three areas (‘C. Owen’). However, the remaining three putative species were not recovered in our Structure analyses. Increasing the number of populations modelled by Structure (i.e. the value of K) did not result in subdivision of the other collections in line with the hypothesised species based on morphology, except at one of the three nearby collecting sites (near Staircase, ). AFLP profiles for the Marino Mountains populations were also able to distinguish ‘C. Owen’ from the other Marino Mountains samples, but provided no clear signal for groupings among the others (I Breitwieser et al., unpubl.). Moreover, our Structure results cast doubt on whether any of the plants at the Marino Mountains are conspecific with ‘C. elongata’ or ‘C. long hairs’ from Mt Arthur. However, in a neighbour-joining tree one of the populations from the Marino Mountains (that composed of plants collected as ‘C. elongata’, ‘C. calcicole’ and ‘C. long hairs’) inferred using Structure did group with ‘C. elongata’ from Mt Arthur (), so a relationship between them should not be entirely ruled out.
Reconciling our SSR data with morphology for these Marino Mountains plants will require reappraisal of morphological variation at the site, but one possibility is that a hybrid swarm has formed, perhaps involving ‘C. elongata’ as one parent. Allan (Citation1961) suggested that there was good evidence of hybridism among forms of Craspedia and that introgression may occur, but specific testable hypotheses are lacking. Another possibility is that morphological variation in Marino Mountains Craspedia is partly the result of a phenotypically plastic species. This will be difficult to resolve until biological species have been discriminated in New Zealand Craspedia more generally. A priority for future research should be the examination of widespread morphological forms for genetic cohesion and hybridisation with morphologically distinct sympatric populations over their full range.
In summary, we have characterised a set of 12 polymorphic SSR markers for New Zealand Craspedia that can be used to identify and relate genetic populations. Assaying these markers in samples from three mountain areas of Kahurangi National Park supports the contention that numerous biological species occur there because at least some morphologically distinguishable forms are genetically distinct and persist in sympatry. However, hybridisation may occur at some sites between some genetic groups, but not between other groups or at other sites, and crossing relationships may be complex in the genus as a whole in New Zealand. Our results support the genetic cohesion of some of Druce's (Citation1993) informally named putative species of Kahurangi National Park that are based on morphology with emphasis on leaf characters (‘C. long hairs’ at Mt Arthur and Mt Mytton, ‘C. marble’ at Mt Arthur = ‘C. Mytton’ at Mt Mytton, ‘C. Owen’ on the Marino Mountains), but cast doubt on others. However, formal taxonomic recognition of these putative species would be premature without the consideration of more populations of Craspedia from Kahurangi National Park and throughout New Zealand because their circumscription remains problematic.
Acknowledgements
We thank Tim Galloway for assistance with fieldwork and Heidi Meudt, Peter Heenan and Christine Bezar for comments on the draft manuscript. Two anonymous referees also provided constructive criticism. RNA-seq was carried out by Duckchul Park (Landcare Research). The research was supported by Core funding for Crown Research Institutes from the Ministry of Business, Innovation and Employment's Science and Innovation Group.
References
- Allan HH 1961. Flora of New Zealand. Vol. 1. Wellington, Government Printer.
- Botstein D, White RL, Skolnick M, Davis RW 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32: 314–331.
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF 2001. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques 31: 24–28.
- Breitwieser I, Ford KA, Smissen RD 2010. A test of reproductive isolation among three sympatric putative species of Craspedia (Asteraceae: Gnaphalieae) at Mt Arthur in New Zealand. New Zealand Journal of Botany 48: 75–81.
- Dawson MI 1995. Contributions to a chromosome atlas of the New Zealand flora—33. Miscellaneous species. New Zealand Journal of Botany 33: 477–487.
- Dawson MI, Breitwieser I, Ward JM 1999. Chromosome numbers in Craspedia, Ewartia and Pterygopappus (Compositae—Gnaphalieae). Australian Systematic Botany 12: 671–674.
- Doyle JJ, Dickson EE 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon 36: 715–722.
- Druce AP 1993. Indigenous vascular plants of New Zealand (9th revision). Lincoln, Unpublished checklist held at Landcare Research Library.
- Druce AP, Williams PA, Heine JC 1987. Vegetation and flora of tertiary calcareous rocks in the mountains of western Nelson, New Zealand. New Zealand Journal of Botany 25: 41–78.
- Earl DA, von Holdt BM 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4: 359–361.
- Evanno G, Regnaut S, Goudet J 2005. Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology 14: 2611–2620.
- Faircloth BC 2008. MSATCOMMANDER: detection of microsatellite repeat arrays and automated, locus-specific primer design. Molecular Ecology Resources 8: 92–94.
- Falush D, Stephens M, Pritchard JK 2003. Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
- Ford KA, Ward JM, Smissen RD, Wagstaff SJ, Breitwieser I 2007. Phylogeny and biogeography of Craspedia (Asteraceae: Gnaphalieae) based on ITS, ETS and psbA-trnH sequence data. Taxon 56: 783–794.
- Lee D, Nelson C 2008. Limehouse rock. In: Graham IJ ed. A continent on the move: New Zealand Geoscience into the 21st century. Wellington, GSNZ Miscellaneous Publication 124. 388 p.
- Liu K, Muse SV 2005. Powermarker: integrated analysis environment for genetic marker data. Bioinformatics 21: 2128–2129.
- Nei M 1972. Genetic distance between populations. The American Naturalist 106: 283–292.
- Nybom H, Weising K, Rotter B 2014. DNA fingerprinting in botany: past, present, future. Investigative Genetics 5: 1.
- Pritchard JK, Stephens M, Donnelly P 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
- Pritchard JK, Wen X, Falush D 2010. Documentation for structure software: version 2.3. http://pritch.bsd.uchicago.edu/structure.html (accessed 2 February 2014).
- Rogers G, Walker S 2005. Evolution of the New Zealand vascular flora: regional and provincial patterns of richness, radiation, and endemism. New Zealand Journal of Botany 43: 381–414.
- Rozen S, Skaletsky HJ 2000. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S eds. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ, Humana Press. Pp. 365–386.
- Saitou N, Nei M 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425.
- Smissen RD, Heenan PB 2007. DNA fingerprinting supports hybridisation as a factor explaining complex natural variation in Phormium (Hemerocallidaceae). New Zealand Journal of Botany 45: 419–432.
- Vos P, Hogers R, Bleeker M, Reijans M, van der Lee T, Hornes M 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23: 4407–4014.