ABSTRACT
We examined the origins of Sophora seeds collected from the beaches of the Kermadec and Chatham Islands using chloroplast DNA sequencing and comparison with published sequences of known provenance. Sophora does not grow on the Kermadec Islands but two species grow on the Chatham Islands, both in low numbers. All seeds sequenced were confirmed to be from Sophora sect. Edwardsia. Several seeds exhibited novel DNA sequences that had not been found in past sampling. The remaining haplotypes have all been previously detected in the North and/or South Islands and some have also been recorded from Chile, Gough Island or the Juan Fernandez Islands. The two haplotypes that had been sequenced in the resident Chatham Islands S. chathamica population were both found in the beach-cast seeds from the islands. However, an additional four haplotypes were also detected in the Chatham Islands beach-cast seeds, indicating dispersal from elsewhere.
Introduction
Oceanographic features, such as currents, can strongly influence the diversity patterns of taxa with sea-dispersed propagules (Gillespie et al. Citation2012). They provide directionality and can either promote gene flow or act as a barrier to dispersal. Beach-cast biological specimens have the potential to contribute to our understanding of oceanic connectivity patterns but have only been examined in a few marine taxa (e.g. Collins et al. Citation2010; Bussolini & Waters Citation2015).
Sophora sect. Edwardsia Salisb. is well known for its Southern Hemisphere distribution. Phylogenetic analyses have indicated that the 19 species in the section are closely related and have a recent origin (Hurr et al. Citation1999; Mitchell & Heenan Citation2002; Shepherd & Heenan Citationsubmitted). Sister to sect. Edwardsia, and the type species for the genus Sophora (Heenan et al. Citation2004), is S. tomentosa in sect. Sophora L., a pantropical coloniser of beaches and sand dunes in Africa, Asia and central America. It is well known for the dispersal of its seeds by ocean currents (e.g. Ridley Citation1930). The distribution of Sophora sect. Edwardsia is also thought to result from oceanic dispersal of its buoyant seeds (Heenan et al. Citation2001). Charles Darwin used the genus to support his idea that the similarity of the New Zealand and South American floras resulted from seed dispersal across oceans, suggesting that Sophora seeds may have floated between New Zealand and Chile (Darwin Citation1887). The seeds have a hard and impervious seed coat protecting the embryo and can remain afloat and viable in seawater for years (Sykes & Godley Citation1968). Sophora seed dispersal may also occur through the ingestion by seabirds of seeds floating at sea, followed by regurgitation (Tennyson Citation1995; Smith Citation2012). In a phylogenetic study of the section, DNA haplotypes of Sophora were found to be shared between Chile, New Zealand, Gough Island and Juan Fernandez Island, indicating recent connections (Shepherd & Heenan submitted). In contrast, islands further north in the Pacific (Hawaii, French Polynesia, Easter Island and Lord Howe Island) and Réunion Island in the Indian Ocean each exhibited unique haplotypes suggesting that the species on these islands are more isolated.
Within New Zealand, which with eight species is the centre of species diversity for S. sect. Edwardsia, a chloroplast phylogeographic study indicated moderate population differentiation but found variation was not partitioned by species boundaries (Shepherd et al. Citation2017) and therefore species could not be identified by chloroplast sequences. Although some haplotypes had wide distributions many others were restricted to small areas.
Here we examine the origins of beach-cast Sophora seeds collected from the Chatham and Kermadec Islands. The Chatham Islands archipelago, 850 km east of the main New Zealand islands (considered here as the North, South and Stewart Islands), does have several small populations of S. chathamica on Chatham Island. The largest and most well-known population is located on the margins of Te Whanga Lagoon, which is often closed off from the sea (Molloy Citation2002). Past sequencing of individuals from this population revealed two chloroplast haplotypes, both of which also occurred in mainland New Zealand (Shepherd et al. Citation2017).
Sophora seeds, which are a distinctive bright yellow colour, are a common sight on Chatham Island beaches. For example, Molloy (Citation2002) collected 256 seeds over a 400 m stretch of beach in a single day. Scientists have speculated about the origins of these seeds for over 150 years. Travers (Citation1865) found a Sophora seed on the northwest coast of Pitt Island, which he assumed had floated on currents from mainland New Zealand. He also noted the presence of wood from mainland New Zealand trees on Chatham Island beaches, which he concluded indicated the existence of direct oceanic currents from mainland New Zealand to the Chatham Islands. Cockayne (Citation1902) disputed Travers’ mainland New Zealand origin hypothesis for these beach-cast Sophora seeds and suggested they more likely to come from the resident Chatham Island population of S. chathamica. Molloy (Citation2002) germinated Sophora seeds collected from the strandline of Te One Beach on Chatham Island. He reported that the majority of resulting seedlings had the divaricating morphology characteristic of the most common mainland New Zealand species S. microphylla, rather than the non-divaricating seedling form of resident S. chathamica, supporting a mainland New Zealand origin. However, more recently, three small S. microphylla trees were found at Blind Jims, on the margin of Te Whanga Lagoon, on Chatham Island (CHR 604802; de Lange et al. Citation2011). The origin of these plants is uncertain as they occur in a site well away from where any beach strand seed could occur and they may derive from cultivated plants used in ecological restoration plantings at the site.
Based on data from the Global Drifter Program, which deploys satellite-tracked drifters at a 15 m depth, and simulations of propagule dispersal, the predominant currents arriving in the Chatham Islands originate from the east coasts of the North and South Islands (Chiswell & Rickard Citation2006, Citation2011). The dispersal time from mainland New Zealand to the Chatham Islands has been estimated to be at least 82 days (Chiswell & Rickard Citation2011).
In contrast to the Chatham Islands, the Kermadec Islands archipelago, which is 920 km north of the North Island, does not have a resident population of Sophora. Sykes & Godley (Citation1968) collected Sophora seed from the beaches of Raoul and Macauley Islands in the Kermadec Islands. They suggested these seeds originated from mainland New Zealand and, like Travers, used the presence of wood from endemic New Zealand trees recorded on the shores of the Kermadec Islands (Sykes & Godley Citation1968; Oliver Citation1910) as support for dispersal from mainland New Zealand. However, seeds of tropical plant species have also been recorded on Kermadec Island beaches (Sykes & Godley Citation1968), indicating dispersal can also occur from the north. The Kermadecs are situated near the centre of the South Pacific Subtropical Gyre, which is characterised by weak, variable currents (Sutton et al. Citation2012). Dispersal simulations have indicated that drift dispersal between the mainland and the Kermadecs takes 30–50 days (Sutton et al. Citation2012).
Our main aim was to use chloroplast DNA sequences to examine the origins of Sophora seeds collected from beaches of the Kermadec and Chatham Islands. In particular we wanted to test whether the beach-cast seeds from the Chatham Islands shared haplotypes with the resident S. chathamica population.
Methods
Leaves from seedlings germinated from 13 Chatham Islands beach-cast seeds that had been collected by Molloy (Citation2002), and from four Kermadec Islands beach-cast seeds collected by Sykes & Godley (Citation1968), were dried in silica gel. Genomic DNA was obtained from these silica gel-dried samples using a modified-CTAB extraction method (steps 1, 3–7 from table 1 in Shepherd & McLay Citation2011). Polymerase chain reaction and sequencing of the chloroplast trnQ-5′rps16 and trnHGUG -psbA intergenic spacers were performed as described in Shepherd & Heenan (submitted).
Two datasets were constructed. The first, a full-length dataset where the DNA sequences for both chloroplast loci from the seeds were aligned with the sequences from Shepherd & Heenan (submitted). These sequences represent the diversity found within mainland New Zealand and also include sequences from all other members of Sophora sect. Edwardsia, except S. masafuerana. In the second dataset the trnQ-5′rps16 sequences from the seeds were aligned with the sequences of 416 known provenance samples of Sophora sect. Edwardsia from mainland New Zealand (Shepherd et al. Citation2017). For the trnQ-5′rps16 haplotypes sequenced from the seeds, this larger sample set allowed comparison with the distribution and frequency of the same haplotypes found in mainland New Zealand and the resident Chatham Island S. chathamica.
For both datasets, sequences were aligned with MUSCLE (Edgar Citation2004) with default parameters and manual alignment of large indels (insertions and deletions). For alignment 2, indels in mononucleotide runs were recoded as single events. For alignment 1, mononucleotide repeats of varying length were excluded, since analyses in Shepherd & Heenan (submitted) indicated reticulation in this dataset and these regions are prone to homoplasy at larger geographic scales (Ingvarsson et al. Citation2003). For both datasets the remaining indels, and a 6 bp inversion in the trnHGUG -psbA intergenic spacer in alignment 1, were recoded as single events.
Median-joining networks (Bandelt et al. Citation1999) were produced for each alignment using Network v5.0.0.0 (www.fluxus-engineering.com) and transitions and transversions were equally weighted.
Results
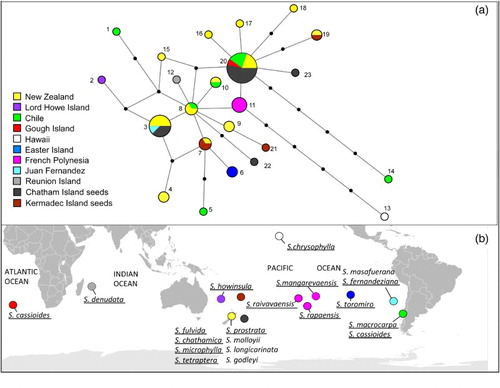
All newly-generated DNA sequences have been deposited in GenBank (accession numbers KY555093–KY555126). Alignment 1 had a total of length of 908 bp (552 bp and 356 bp for the trnQ-5′rps16 and trnHGUG -psbA intergenic spacers, respectively) prior to indel recoding and exclusion of mononucleotide runs. Following the recoding of indels and removal of mononucleotide runs, 23 haplotypes were detected (named 1 to 23). The relationships between them are shown in the median-joining network (). Seven of the 23 haplotypes were detected in the beach-cast seeds (three from the Kermadec Islands and four from the Chatham Islands). Three haplotypes were only detected from beach-cast seeds (one from the Kermadec Islands and two from the Chatham Islands) and each differed by one to two mutational changes from its closest related haplotype (). The remaining three Kermadec Islands seeds possessed haplotypes only detected from New Zealand (one seed had haplotype 19 and two had haplotype 7). Eight of the Chatham Islands seeds had haplotype 20, which was also found in New Zealand, Chile and Gough Island in the Atlantic Ocean. Another three seeds had haplotype 3, which occurs in New Zealand and the Juan Fernandez Islands.
Figure 1. A, Median-joining network of alignment 1 dataset, which was the trnQ-5′rps16 and trnHGUG -psbA intergenic spacers, from Sophora sect. Edwardsia. Circle size is proportional to frequency and small black circles represent missing intermediate haplotypes; B, the distribution of S. sect. Edwardsia. Species included in the sampling for alignment 1 are underlined. The locations of the Chatham and Kermadec Islands are also indicated.
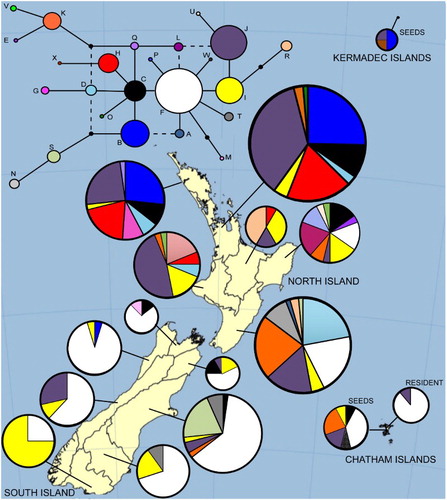
For alignment 2, which comprised only the trnQ-5′rps16 region with mononucleotide repeat length changes coded, a total of 24 haplotypes were defined (named A to X). Eight haplotypes were detected from the beach-cast seeds (three from the Kermadec Islands and six from the Chatham Islands). Two of these haplotypes had not been found in our previous sampling from New Zealand. One of these, haplotype W from the Chatham Islands, was most closely related to haplotype F, from which it differed by a single base pair substitution (). The other unique haplotype, haplotype X, from the Kermadec Islands, also differed by a 1 bp substitution from its most closely related haplotype, haplotype H (). Of the remaining three Kermadec Islands seeds, two had haplotype B and one had haplotype J. Haplotype B is most common in the northern North Island (). Haplotype J has a widespread distribution in New Zealand but is more common in the North Island, particularly in the north and west (). Two seeds from the Chatham Islands also had haplotype J. This haplotype has also been detected from the resident S. chathamica population on the Chatham Islands. The most common haplotype in the remaining Chatham Islands seeds was haplotype F (detected in five seeds). This haplotype is the most common haplotype in the South Island, is also found in the southern North Island and is the most common haplotype in the resident Chatham Islands S. chathamica population. Three Chatham Islands seeds contained haplotype K. This haplotype is most frequent in the eastern North Island but has been detected once in the eastern South Island. Finally, haplotypes C and I were each detected from a single Chatham Islands seed. Both these haplotypes have widespread distributions in New Zealand but haplotype C is uncommon in the South Island.
Figure 2. Median-joining network of alignment 2 sequences (the trnQ-5′rps16 intergenic spacer). The haplotypes present in each Ecological Province of New Zealand are indicated (precise sample location is given in Shepherd et al. Citation2017). Pie sizes are proportional to frequency.
Discussion
A high diversity of haplotypes was detected, even with our relatively small sampling of Sophora beach-cast seeds. Several of these haplotypes had not been detected in our previous sampling of S. sect. Edwardsia from the main New Zealand islands or overseas (Shepherd et al. Citation2017; Shepherd & Heenan submitted), suggesting diversity remains to be found within the section. Our previous study of 416 Sophora individuals from New Zealand (Shepherd et al. Citation2017) found that rare haplotypes were a feature of the New Zealand dataset with 15 of the 22 haplotypes occurring at a frequency of less than 5% and six of these are only found in one to two individuals. Although it is not possible to determine the exact origins of these novel beach-cast haplotypes, it is notable that each of them was most closely related to haplotypes detected from New Zealand (although sampling effort was also considerably greater in New Zealand than elsewhere).
For the seeds that exhibited haplotypes that had been detected in our previous sampling, no haplotypes were shared with the north Pacific Islands (Hawaii, French Polynesia, Lord Howe Island or Easter Island) or Réunion Island in the Indian Ocean. Instead all remaining seeds shared haplotypes that have been sequenced from New Zealand and, for some haplotypes, also sequenced from Chile and Gough Islands or the Juan Fernandez Islands. Given the predominant current patterns it is possible that all the beach-cast seeds sampled from the Kermadec and Chatham Islands originated from New Zealand, but a South American origin for some of the seeds cannot be excluded based on our data.
For alignment 2, which compared the beach-cast seed trnQ-5′rps16 sequences with an extensive database of New Zealand sequences, apart from the novel haplotypes, the haplotypes found in the beach-cast seeds were reasonably common and/or had widespread distributions on the New Zealand mainland. This limited their usefulness for determining beach-cast seeds origins.
Six haplotypes were detected from the Chatham Islands seeds for alignment 2. Only two of these haplotypes had been sequenced from the resident Chatham Islands S. chathamica population (haplotype F and J); these haplotypes were also common on mainland New Zealand. The remaining four haplotypes are likely to be a result of dispersal, supporting the suggestion of seed dispersal from New Zealand to the Chatham Islands (Travers Citation1865; Molloy Citation2002).
In conclusion, the genetic analysis of beach-cast Sophora seeds has provided some insights into seed origins, with many of the seeds examined likely dispersing from the main New Zealand islands to more distant offshore islands. However, the limited genetic structuring observed in the section and the wide distributions of many haplotypes, both within mainland New Zealand and across the southern Pacific Ocean, limits provenance determination of dispersed seeds. This result is comparable to previous studies of the genetic relationships of Chatham Islands plants. Heenan et al. (Citation2010) showed that a considerable number of species endemic to the Chatham Islands have as their closest relative common and widespread species from the main New Zealand islands. In a similar way, the fern Asplenium hookerianum has two haplotypes on the Chatham Islands that are more closely related to haplotypes from the main New Zealand islands than they are to each other (Shepherd et al. Citation2009).
Acknowledgements
We thank Bill Sykes and Brian Molloy for astutely collecting seeds from the Kermadec Islands and Chatham Islands, respectively. Associate Editor: Dr Peter de Lange.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bandelt H-J, Forster P, Rohl A. 1999. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036
- Bussolini LT, Waters JM. 2015. Genetic analyses of rafted macroalgae reveal regional oceanographic connectivity patterns. Journal of Biogeography. 42:1319–1326. doi: 10.1111/jbi.12491
- Chiswell SM, Rickard GJ. 2006. Comparison of model and observational ocean circulation climatologies for the New Zealand region. Journal of Geophysical Research. 111:C10011, doi: 10.1029/2006JC003489
- Chiswell SM, Rickard GJ. 2011. Larval connectivity of harbours via ocean currents: a New Zealand study. Continental Shelf Research. 31:1057–1074. doi: 10.1016/j.csr.2011.03.012
- Cockayne L. 1902. A short account of the plant-covering of Chatham Island. Transactions and Proceedings of the New Zealand Institute. 34:243–325.
- Collins CJ, Fraser CI, Ashcroft A, Waters JM. 2010. Asymmetric dispersal of southern bull-kelp (Durvillaea antarctica) adults in coastal New Zealand: testing an oceanographic hypothesis. Molecular Ecology. 19:4572–4580. doi: 10.1111/j.1365-294X.2010.04842.x
- Darwin F, ed. 1887. The life and letters of Charles Darwin, including an autobiographical chapter. Vol 2. London: John Murray.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32:1792–1797. doi: 10.1093/nar/gkh340
- Gillespie RG, Baldwin BG, Waters JM, Fraser CI, Nikula R, Roderick GK. 2012. Long-distance dispersal: a framework for hypothesis testing. Trends in Ecology & Evolution. 27:47–56. doi: 10.1016/j.tree.2011.08.009
- Heenan PB, Dawson MI, Wagstaff SJ. 2004. The relationship of Sophora sect. Edwardsia (Fabaceae) to Sophora tomentosa, the type species of the genus Sophora, observed from DNA sequence data and morphological characters. Botanical Journal of the Linnean Society. 146:439–446. doi: 10.1111/j.1095-8339.2004.00348.x
- Heenan PB, de Lange PJ, Wilton AD. 2001. Sophora (Fabaceae) in New Zealand: taxonomy, distribution, and biogeography. New Zealand Journal of Botany. 39:17–53. doi: 10.1080/0028825X.2001.9512715
- Heenan PB, Mitchell, AD, de Lange PJ, Keeling J, Paterson AD 2010. Late-Cenozoic origin and diversification of Chatham Islands endemic plant species revealed by analyses of DNA sequence data. New Zealand Journal of Botany. 48:83–136. doi: 10.1080/0028825X.2010.494337
- Hurr KA, Lockhart PJ, Heenan PB, Penny D. 1999. Evidence for the recent dispersal of Sophora (Leguminosae) around the Southern Oceans: molecular data. Journal of Biogeography. 26:565–577. doi: 10.1046/j.1365-2699.1999.00302.x
- Ingvarsson PK, Ribstein S, Taylor DR. 2003. Molecular evolution of insertions and deletion in the chloroplast genome of silene. Molecular Biology and Evolution. 20:1737–1740. doi: 10.1093/molbev/msg163
- de Lange PJ, Heenan PB, Rolfe JR. 2011. Checklist of vascular plants recorded from the Chatham Islands. Wellington: Department of Conservation.
- Mitchell AD, Heenan PB. 2002. Sophora sect. Edwardsia (Fabaceae): further evidence from nrDNA sequence data of a recent and rapid radiation around the Southern Oceans. Botanical Journal of the Linnean Society. 140:435–441. doi: 10.1046/j.1095-8339.2002.00101.x
- Molloy BPJ. 2002. Origin of Sophora chathamica (Fabaceae) on Chatham Island. Canterbury Botanical Society. 36:37–46.
- Oliver WRB. 1910. The vegetation of the Kermadec Islands. Transactions and Proceedings of the New Zealand Institute. 42:118–175.
- Ridley HN. 1930. The dispersal of plants throughout the world. Kent: Reeve.
- Shepherd LD, de Lange PJ, Perrie LR. 2009. Multiple colonizations of a remote oceanic archipelago by one species: how common is long-distance dispersal? Journal of Biogeography. 36:1972–1977. doi: 10.1111/j.1365-2699.2009.02120.x
- Shepherd LD, de Lange PJ, Perrie LR, Heenan PB. 2017. Chloroplast phylogeography of New Zealand Sophora trees (Fabaceae): extensive hybridization and widespread Last Glacial Maximum survival. Journal of Biogeography. doi:10.1111/jbi.12963.
- Shepherd LD, Heenan PB. Submitted. Molecular phylogeny of Sophora sect Edwardsia (Fabaceae). New Zealand Journal of Botany.
- Shepherd LD, McLay TGB. 2011. Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. Journal of Plant Research. 124:311–314. doi: 10.1007/s10265-010-0379-5
- Smith JMB. 2012. Evidence for long-distance dispersal of Sophora microphylla to sub-Antarctic Macquarie Island. New Zealand Journal of Botany. 50:83–85. doi: 10.1080/0028825X.2011.621715
- Sutton P, Chiswell S, Gorman R, Kennan S, Tickard G. 2012. Physical marine environment of the Kermadec Islands region. Science for Conservation. 318, Department of Conservation, Wellington.
- Sykes WR, Godley EG. 1968. Transoceanic dispersal in Sophora and other genera. Nature. 218:49–496. doi: 10.1038/218495a0
- Tennyson AJD. 1995. Flora of Karewa Island, Bay of Plenty. Tane. 35:17–23.
- Travers HH. 1865. Notes on the Chatham Islands (lat. 44° 30’ S. long. 175 W.). Journal of the Linnean Society of London, Botany. 9:135–144. doi: 10.1111/j.1095-8339.1865.tb00017.x
