Abstract
Clostridioides difficile infection may be complicated by co-infection with other pathogens. We here describe the successful use of faecal microbiota transplantation to eradicate concomitant C. difficile and extensively drug-resistant (XDR) KPC-producing Klebsiella pneumoniae. Donor microbiota efficiently engrafted in the patient, and a donor-like microbial assemblage persisted in the patient during six months follow-up. The report explores the potential for the donor microbiota to eradicate and replace multi-resistant microorganisms.
Introduction
Faecal microbiota transplantation (FMT) is an emerging therapeutic option to restore a deteriorated intestinal microbiota [Citation1]. It involves the transfer of processed feces from a healthy donor to a recipient and is now an established and highly efficient treatment for recurrent Clostridioides difficile infection (CDI) with cure rates above 90% [Citation2,Citation3].
Antimicrobial resistance is an imminent global health threat, and the continued emergence and spread of multidrug-resistant bacteria seriously reduce viable treatment options [Citation4]. Asymptomatic colonization of multidrug-resistant bacteria is common, and the gut serves as a natural reservoir that may acquire and share antimicrobial resistance through horizontal gene transfer [Citation5]. Carbapenem-resistant Enterobacterales such as Klebsiella pneumoniae and Escherichia coli constitute a particular challenge due to their propensity to cause fatal infections [Citation4]. Intestinal decolonization of antibiotic-resistant strains remains a challenge, and recent studies indicate the potential of FMT to promote competitive exclusion by introducing an ecologically well-balanced and complex microbial population [Citation6].
We here report the use of FMT to eradicate a severe, refractory CDI complicated by co-carriage with XDR K. pneumoniae, susceptible only to ceftazidime-avibactam. We further explore the potential for the donor microbiota to eradicate and replace multi-resistant microorganisms.
Methods
The patient provided written consent for participation and reporting of the case. Antimicrobial susceptibility testing for all antibiotics except fosfomycin was performed using freeze-dried broth microdilution plates (Sensititre, TREK Diagnostic System, Cleveland, OH) according to the manufacturer’s recommendations. Antimicrobial susceptibility testing for fosfomycin was carried out by Etest (BioMerieux, Marcy-l’Étoile, France) according to the manufacturer’s instructions. Total bacterial community DNA was extracted from faecal samples (29–246 mg) obtained from the patient on Days -6, 14, 56, 105, 168 and 203 relative to the FMT, as well as from the donor sample, by use of DNeasy PowerLyzer PowerSoil Kit (Qiagen) and the concentration determined fluorometrically (Qubit™ dsDNA HS Assay, ThermoFisher Scientific) according to supplier recommendations. Total bacterial load was determined in faecal samples by quantitative PCR (qPCR) targeting the 16S rRNA gene as previously described [Citation7]. To profile the bacterial community composition, 5 ng DNA was used as template for Ion Torrent sequence library preparation, based on the V3 hypervariable region, as previously described [Citation8]. Partial 16S rRNA gene sequencing was subsequently performed on an Ion Personal Genome Machine® (PGM™, ThermoFisher Scientific) using Ion PGM Hi-Q kit, 200 bp sequencing and Ion 318™ Chip. Bioinformatic processing was performed essentially as previously described [Citation8]. Briefly, raw FASTQ sequence data were initially processed in CLC Genomic workbench (Version 12, Qiagen) to de-multiplex samples and trim reads for sequencing primers. Next, the DADA2 pipeline (version 1.12.1) [Citation9] implemented in RStudio (RStudio Inc, Boston, MA) was used to generate an amplicon sequence variant (ASV) table (MaxEE = 2, pool = TRUE) and taxonomic classification of the inferred ASVs based on the Ribosomal Database Project (rdp_train_set_16) [Citation10]. QIIME2 was used for downstream processing of the ASV table [Citation11]. The median number of high-quality reads for the seven included samples was 61,162 (40,206–81,307) which represented a total of 327 features (ASVs). The qiime diversity core-metrics-phylogenetic script with a sampling depth of 40,000 was run to determine alpha- and beta diversity as well as taxonomical composition in the samples. The 16S rRNA gene sequence data are deposited in the NCBI Sequence Read Archive with the accession number PRJNA562138.
Case presentation
A 69 years-old woman was referred with severe recurrent CDI, refractory to antimicrobial treatment and complicated by intestinal co-colonization with a KPC-producing, XDR K. pneumoniae. She had severe comorbidity with atherosclerosis, heart failure, atrial fibrillation, and previous episodes of ventricular fibrillation, necessitating a dual chamber cardioverter-defibrillator (ICD) and anticoagulant therapy. Prior to referral, she had suffered warfarin-induced intestinal bleeding and been admitted for intensive care and coagulopathy treatment. She had received piperacillin-tazobactam for a urinary tract infection with sepsis. On discharge, she had persistent diarrhea which exaggerated the coagulopathy. She improved with intravenous fluid replacement, but diarrhea and abdominal pain persisted alongside marked leukocytosis (21.1 per 10−9 L) indicative of severe C. difficile enteritis. Stool tests were qPCR positive for C. difficile toxin A/B and binary toxin. Vancomycin 125 mg QID induced clinical and biochemical improvement, but diarrhea recurred five days after cessation.
At readmission three weeks later, the patient was dehydrated and fatigued with fever, abdominal pain, and more than 20 bowel openings per day. She had leukocytosis and fluid derangement, compatible with severe disease. After 5 days fluid replacement, she was discharged with four weeks tapered vancomycin. This was insufficient in preventing CDI recurrence, and she was readmitted severely dehydrated and somnolent. Because improvement with vancomycin had been negligible, she was started on fidaxomicin. Surveillance cultures revealed XDR K. pneumoniae from feces (). In compliance with local guidelines, she was placed in contact isolation. Her diarrhea persisted despite fidaxomicin, and given the burden of prolonged admission in contact isolation, she was referred for FMT as a last resort.
Table 1. Antimicrobial susceptibility of a XDRKPC-producing Klebsiella pneumoniae, eradicated from the gut using faecal microbiota transplantation. Minimum inhibitory concentrations are interpreted in accordance with CLSI susceptibility criteria.
At the initial assessment, the patient’s medical history and current medications were carefully reviewed in order to reduce the risk of recurrence, with particular emphasis on proton pump inhibitor (which she did not receive) and other antibiotics (which had been stopped). She consented to FMT. The FMT was performed after 12 days fidaxomicin pre-treatment and by colonoscopy, using fresh donor material from a healthy, unrelated, anonymous donor who had been thoroughly screened according to a standard protocol and who had been used as a donor to treat other patients with recurrent CDI [Citation12]. The donor material consisted of 72 g of faeces which had been homogenized and suspended in 500 ml of isotonic sodium chloride, and the suspension was evenly distributed through the working channel of the colonoscope into the coecum and ascending colon. A prompt response followed the FMT with resolution of diarrhea and marked improvement in general well-being. One week after the FMT, faecal CD toxin test and culture for carbapenem-resistant organisms were negative. Sustained clearance was confirmed, and the patient remained stable.
Results
Bacterial load and community composition in patient and donor
A marked increase in both bacterial load and diversity was observed following FMT and remained high for at least 203 days (). The bacterial composition in the patient before the FMT (Day -6) was characterized by low bacterial richness comprising only 40 amplicon sequence variants (ASVs), which was 6.5-fold less than observed in the donor. The four most dominant ASVs, which together constituted 72% of the community on Day -6, were classified as Clostridium ramosum, Clostridium citroniae, Erysipelotrichaceae and Enterobacteriaceae (). Five different ASVs classified as Klebsiella spp. were identified in the patient on Day -6, collectively representing 8% of the community, while 0.2% were classified as K. pneumonia. We did not detect C. difficile nor the family Peptostreptococcaceae (Clostridium cluster XI) containing this species on Day -6.
Figure 1. (A) Composition of the gut microbiota in donor and patient before and after FMT. Stacked relative abundances of amplicon sequence variants (ASVs) constituting a minimum 2% of the total community in any sample are shown and colored either according to taxonomy (phylum level) or otherwise highlighted as indicated (Gray: ASVs found in patients prior to FMT, red: Proteobacteria, blue: Firmicutes, green: Bacteroidetes, black: aggregated ASVs constituting <2% of the community). Percentages indicating the fraction of ASVs found in the donor, which were also detected in the patient sample, as well as the fraction of ASVs found in the patient sample, which were not detectable in the donor, are indicated for each sample. (B) The total bacterial load estimated as number of 16S rRNA genes (circles) and Shannon diversity index calculated from ASV relative abundances (squares) are shown for patient and donor. (C–E) Plots show relative abundances of ASVs present in donor and patient on the indicated days (Each dot represents one ASV). Spearman correlations are calculated and r-values as well as p-values indicated.
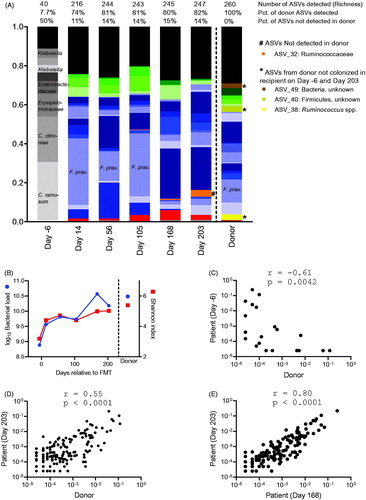
On Day 14 after FMT, 74% of the ASVs identified in the donor were also detectable in the patient. On Day 203, this fraction had increased to 82%, suggesting efficient and persistent engraftment of the donor microbiota (). Approximately, 14% of the ASVs detected in the patient on Day 203 were not found in the donor (). Before FMT, the relative abundances of ASVs identified in both patient and donor were negatively correlated (). After FMT, this switched to a strong positive correlation indicating that the structural microbiota composition was transferred from the donor to the patient (). Heatmap visualization (Supplementary Figure 1) emphasized that after FMT, the patient’s microbiota resembled that of the donor not only by the presence of the donor ASVs, but also by their relative abundances. The correlation between donor and patient on Day 203 was only slightly weaker than within the patient on Day 168 and Day 203 ().
Antimicrobial susceptibility testing
Minimum inhibitory concentrations to 24 antimicrobial agents were determined and interpreted in accordance with CLSI susceptibility criteria (). Due to the lack of a CLSI tigecycline breakpoint for Enterobacterales, no susceptibility categorization could be made for tigecycline, but the isolate had an elevated MIC of 2 mg/L. The K. pneumoniae isolate was resistant to all other antibiotics tested at the time. Subsequent susceptibility testing for ceftazidime-avibactam, which was not available in Denmark in 2015, revealed an MIC of 8 mg/L which is equal to the CLSI susceptibility breakpoint. Susceptibility testing for meropenem-vaborbactam was not performed as it is not available in Denmark.
Discussion
This case report demonstrates the clinical potential of a single FMT to eradicate a severe, refractory CDI complicated by co-carriage of a KPC-producing, XDR K. pneumoniae. It further documents a sustained engraftment of donor microbiota.
After FMT, we observed instant clinical improvement and a rapid increase in microbial community diversity, accompanied by the disappearance of both C. difficile and K. pneumoniae. This suggests short time-to-effect and that eradication of both pathogens was not a time-dependent spontaneous clearance. The finding is consistent with previous studies of FMT for rCDI and reflects de novo establishment of a new microbial ecosystem in the recipient [Citation1].
Neither the species C. difficile nor other species belonging to its family, Peptostreptococcaceae, were detected in the patient before FMT by 16S rRNA amplicon profiling. This was probably due to concomitant fidaxomicin treatment until Day -1, which may have lowered the abundance of C. difficile below the 0.0025% detection level of the sequencing approach. The use of vigorous mechanical and chemical lysis allows the detection of C. difficile spores, but lysis may be incomplete, leading to an underestimation of total C. difficile in the fecal samples. Enterobacterales has previously been found to dominate the microbiota in patients infected with C. difficile prior to FMT treatment [Citation13], and constituted 18% of the microbiota in our patient. The two most predominant ASVs before FMT were anaerobic bacteria belonging to the Firmicutes phylum and classified as C. citroniae (Clostridium XlVa genus) and C. ramosum (Clostridium XVIII genus).
The FMT resulted in almost complete replacement of the microbial community. Up to 82% (Day 203) of the donor ASVs were identified in the patient after FMT compared to only 8% before FMT. One ASV, for which the abundance increased dramatically following FMT, was the butyrate-producing Faecalibacterium prausnitzii (). This species has been associated with treatment success, and a mechanism for butyrate-induced mitigation of C. difficile pathogenicity was recently demonstrated. Interestingly, the F. prausnitzii ASV dominated until Day 105 (34%) and then decreased in abundance until Day 203 (3%). This indicates a dynamic change in recipient microbiota beyond the commonly used eight week follow-up time.
Not only the presence of specific bacterial species (ASVs), but also their relative abundance profile was apparently transferred to the patient and persisted during long-term follow-up. Although we cannot rule out the possibility that some of these ASVs did not originate from the donor the findings in this case study are consistent with reported correlations between faecal communities in donor material and in patients post FMT treatment [Citation13], and further supported by findings that bacterial abundance and phylogeny are the strongest determinants of engraftment based on full metagenomics analysis [Citation14]. Although complete microbiota engraftment is not essential for successful CDI treatment [Citation15], the observed long-term stability of the transferred community assemblage underpins the importance of communal interactions between members of the microbiota to maintain a stable community. This highlights the importance of using material from a single donor with a well-balanced microbiota to ensure both successful treatment of the C. difficile infection and a long-term healthy microbiota in the patient. Additionally, it underlines the importance of screening of potential donors for microbiota-related adverse conditions as well as antimicrobial resistance determinants that might be unintentionally transferred [Citation6].
Our findings have important implications for clinical care. FMT is a viable treatment option to eradicate both C. difficile and multidrug-resistant organisms from the intestine. Our finding of sustained engraftment points to important details in the follow-up of patients who receive FMT. Long-term follow-up, i.e. at least 26 weeks, may provide important insights in microbiota changes that ensure a beneficial stable condition. Diagnostic testing should be multiway and include methods suitable for in-depth identification of low-abundant pathogens. Lastly, co-infection with C. difficile and multidrug-resistant organisms may pose particular diagnostic and therapeutic challenges.
Supplemental Material
Download PDF (38.5 KB)Acknowledgements
The authors thank Bodil Madsen for excellent technical support as well as Marlene Danner Dalgaard and NeslihanBicen at the DTU Multi-Assay Core facility for performing the 16S rRNA sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Allegretti JR, Mullish BH, Kelly C, et al. The evolution of the use of faecal microbiota transplantation and emerging therapeutic indications. Lancet. 2019;394(10196):420–431.
- McDonald LC, Gerding DN, Johnson S, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66(7):987–994.
- Kelly CR, Khoruts A, Staley C, et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent Clostridium difficile infection: a randomized trial. Ann Intern Med. 2016;165(9):609–616.
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281.
- Penders J, Stobberingh EE, Savelkoul PH, et al. The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol. 2013;4:87.
- Huttner BD, de Lastours V, Wassenberg M, et al. A five-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect. 2019;25(7):830–838.
- Tulstrup MV, Christensen EG, Carvalho V, et al. Antibiotic treatment affects intestinal permeability and gut microbial composition in Wistar rats dependent on antibiotic class. PLoS One. 2015;10(12):e0144854.
- Nielsen LN, Roager HM, Casas ME, et al. Glyphosate has limited short-term effects on commensal bacterial community composition in the gut environment due to sufficient aromatic amino acid levels. Environ Pollut. 2018;233:364–376.
- Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583.
- Cole JR, Wang Q, Fish JA, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(D1):D633–42.
- Bolyen E, Rideout JR, Dillon MR, et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. Peer J. 2018;6:e27295v2.
- Jørgensen SMD, Erikstrup C, Dinh KM, et al. Recruitment of feces donors among blood donors: results from an observational cohort study. Gut Microbes. 2018;9:540–550.
- Weingarden A, Gonzalez A, Vazquez-Baeza Y, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3(1):10.
- Smillie CS, Sauk J, Gevers D, et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 2018;23(2):229–240.e5.
- Staley C, Kelly CR, Brandt LJ, et al. Complete microbiota engraftment Is not essential for recovery from recurrent Clostridium difficile infection following fecal microbiota transplantation. MBio. 2016;7(6):e01965–16.
