Abstract
Objectives
Absence of a functional interferon-λ 4 (IFN-λ4) gene (IFNL4) predicts spontaneous resolution of acute hepatitis C virus (HCV) infections in regions with a predominance of genotype 1, whereas variants of the inosine triphosphate pyrophosphatase (ITPase) gene (ITPA) entailing reduced activity associate with increased sustained virologic response rates following some therapeutic regimens. This study aimed at investigating the impact of IFNL4 on acute HCV genotype 2 or 3 infections, and whether ITPase activity influenced outcome.
Materials and Methods
Two hundred and seven people who injected drugs (PWID) with documented anti-HCV seroconversion, and 57 PWID with reinfection with HCV were analyzed regarding IFNL4 (rs368234815 and rs12979860) and ITPA (rs1127354 and rs7270101), and longitudinally followed regarding HCV RNA.
Results
The spontaneous clearance of HCV infection in anti-HCV seronegative PWID was enhanced when IFN-λ4 was absent (44% vs. 20% for IFNL4 TT/TTrs1368234815 and ΔGrs1368234815 respectively, p < .001; OR 3.2) across genotypes 1-3. The proportion lacking IFN-λ4 was further increased following resolution of repeated re-exposure to HCV (74% among re-infected participants who had cleared at least two documented HCV infections). ITPA genetic variants did not independently impact on the outcome, but among males lacking IFN-λ4, reduced ITPase activity markedly augmented the likelihood of resolution (65% vs. 29% for <100% and 100% ITPase activity, p = .006).
Conclusions
Absence of IFN-λ4 entails an enhanced likelihood of spontaneous resolution both following primary acute infection and repeated re-exposure to HCV across genotypes 1-3. Among men lacking IFN-λ4, reduced ITPase activity improved outcome.
Introduction
Approximately 15-40% of exposed individuals spontaneously resolve acute hepatitis C virus (HCV) infection [Citation1]. Homozygous carriage of the C allele in rs12979860 on chromosome 19, in the proximity of the interleukin 28B gene (IL28B) also known as interferon-λ 3 (IFNL3), increases the likelihood of clearance, especially in geographical regions with a predominance of HCV genotype 1 [Citation2,Citation3]. Recently, however, this polymorphism was found to be located in intron 1 in a previously overlooked gene encoding interferon-λ4 (IFNL4), with the C allele in rs12979860 being in strong linkage disequilibrium (LD) with an insertion/deletion polymorphism, rs368234815, in IFNL4 exon 1. Humans are polymorphic for the dinucleotide TT/ΔG allele in rs368234815 [Citation4], with the TT allele resulting in a disruptive frameshift in the coding region of IFNL4, subsequently entailing pseudogenization and abrogated IFN-λ4 production. In contrast, the ancestral wild-type allele ΔGrs368234815, which is present in non-human primates, allows for production of IFN-λ4.
Two SNPs in the inosine triphosphate pyrophosphatase (ITPase) gene (ITPA), resulting in a missense variant in exon 2 (rs1127354, P32T) and a splice-altering single nucleotide polymorphism (SNP) in intron 2 (rs7270101, IVS2) on chromosome 20, have been demonstrated in a genome-wide association study (GWAS) to protect against ribavirin-induced hemolytic anemia during therapy with IFN-α and ribavirin for HCV infection [Citation5,Citation6], with minor allele carriage resulting in reduced ITPase activity [Citation7]. Reduced ITPase activity, present in approximately one third of HCV infected patients, was significantly associated with increased treatment efficacy mediated by reduced relapse risk following interferon-based therapy for HCV genotype 2/3 [Citation8]. Furthermore, among HCV genotype 1 or 3 infected patients with baseline NS5A resistance-associated substitutions (RASs), reduced ITPase activity improved outcome following some direct acting anti-viral (DAA) regimens [Citation9] secondary to a ribavirin-like reduced relapse risk.
The onset of hepatitis C virus (HCV) infection is often challenging to determine due to the frequent absence of symptoms [Citation10]. However, frequent serological and molecular surveillance of high-risk populations can uniquely identify acute HCV infections. Two recent Swedish studies investigated HCV incidence and spontaneous clearance rates of incident HCV in people who inject drugs (PWID). In the study from Southern Sweden (Malmö), antibodies against HCV were detected at study entry in 60%, and the incidence of new HCV infections was 31/100 person years, with spontaneous clearance seen in 32% [Citation11]. In the study from the Stockholm, baseline anti-HCV antibodies were detected in 77% and ongoing infection in 57%. The incidence of HCV infection in HCV seronegative patients was 26/100 person years, and 19/100 person years in participants with evidence of previous exposure (detectable anti-HCV antibodies but undetectable HCV-RNA). Spontaneous clearance was seen in 20% of seronegative patients and in 44% of those with documented previous exposure [Citation12].
As most studies on acute HCV infection have been conducted in areas with a predominance of HCV genotype 1, this study aimed at investigating spontaneous HCV clearance and host genetic variants in IFNL4 and ITPA in a geographic region with prevalence of HCV genotypes 1, 2 and 3.
Materials and methods
Study population
Patients were recruited from two different needle exchange programs (NEPs), i.e., in Malmö and Stockholm, Sweden. Upon entry to the NEP, the initial baseline serum sample was analyzed for serologies for human immunodeficiency virus (HIV), hepatitis A virus (HAV), hepatitis B surface antigen (HBsAg), hepatitis B core antibody (anti-HBc), and anti-HCV serologies. Subsequently, the participants lacking antibodies against respective virus at enrollment were requested to undergo prospective serologically testing for anti-HIV, HBsAg and anti-HCV at three to six months’ intervals. In Stockholm, HCV RNA was quantified among anti-HCV positive participants at enrollment, and those lacking detectable HCV RNA were included in the present study. Participants from both cohorts lacking exposure to HBV at enrollment were offered vaccination. Samples from Malmö cohort were analyzed first, and to validate the findings from this first evaluation, samples from the Stockholm cohort were subsequently analyzed. As this latter cohort was continuously monitored by means of HCV RNA analysis, the Stockholm material allowed for the additional analysis of patients with reinfection after initially spontaneously clearing an HCV infection.
In the present study, participants from both cohorts who were anti-HCV negative at enrollment, as well as anti-HCV positive but HCV-RNA negative at entry (indicative of past spontaneous clearance of previous HCV exposure) from the Stockholm cohort, were eligible for inclusion. One hundred fifty of 186 anti-HCV seroconverters from the Malmö NEP participants had complete series of pre-baseline, baseline and 1 year follow up HCV-RNA testing, and samples from one hundred thirty nine of these 150 individuals were available for host genetic testing, constituting the Malmö study cohort ().
Figure 1. Flowchart of patients in the study. NEP (needle exchange program), PWID (person who inject drugs), anti-HCV seronegative (anti-HCV antibody assay below the cut-off level of detection at inclusion), and anti-HCV seropositive (anti-HCV antibody assay above the cut-off level of detection at inclusion).
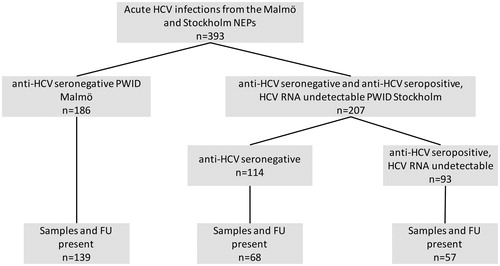
Among the Stockholm NEP participants, 114 anti-HCV negative participants at enrollment who later seroconverted against HCV during the longitudinal study indicating acute HCV infection were identified. Of these 114 patients, 95 samples were available for analyses, and 68 of these patients had had an HCV-RNA test performed >5 months after conversion and were thus included in the study. Ninety-three patients, who had anti-HCV antibodies but HCV RNA undetectable at enrollment (i.e., had resolved at least one prior exposure to HCV), subsequently became re-infected, and of these patients, 57 were available for analysis and had a follow up HCV RNA test >5 months past reinfection (). Five patients from the Stockholm cohort had HIV infection and five had ongoing HBV infection at enrollment.
Definition of spontaneous viral clearance
Spontaneous viral clearance was defined as the absence of viremia in one follow-up sample drawn approximately 12 months after the appearance of anti-HCV antibodies in the Malmö cohort, and in the Stockholm cohort as viral clearance in a sample taken >5 months after either HCV seroconversion or HCV-RNA positivity in patients with previously cleared infections.
HCV-RNA detection and genotyping
All HCV RNA samples were analyzed by Roche COBAS AmpliPrep/COBAS TaqMan HCV Test as described by the manufacturer. All testing from the Malmö cohort was performed at 1/10 dilution in negative serum due to limitations regarding sample volume, hence test sensitivity for these samples was decreased tenfold. Viral genotyping on the first viremic sample in the Malmö cohort was performed by phylogenetic analysis of 321 nucleotides of the NS5B region [Citation13]. HCV genotyping was not assessed in the Stockholm cohort. Information regarding symptoms in conjunction with sampling was not available.
IFNL4 and ITPA genotyping
Two sites were chosen in chromosome 19 (rs12979860 and rs368234815) involved in the expression of IFNL4 and two in chromosome 20 (rs1127354 and rs7270101) known to predict ITPase activity. Briefly: rs1127354 CC and rs7270101 AA corresponds to 100% ITPase activity, CC and AC to 60%, AC and AA to 25%, AC and AC to 10%, and AA and AA to less than 5% respectively [Citation7]. Polymorphisms were determined in serum by allelic discrimination using Taq-Man SNP Assays (Life Technologies) for rs7270101, rs1127354, rs368234815/ss469415590 and for rs12979860 using an in-house test as described previously [Citation14]. As IFNL4 and ITPA are located on chromosomes 19 and 20 respectively, and these genes are unlinked and not in linkage disequilibrium, as chromosomes are segregated independently during meiosis.
Statistical analyses
Chi-Square test or Fisher’s exact test was used to evaluate differences in spontaneous resolution of infection and genotype distribution. One-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test on logarithmic values were used for multiple comparisons of age in the IFNL4 genotype and predicted ITPase activity subgroups. Welch’s T-test on logarithmic values was used when comparing age between two groups of patients. Statistics was done in Prism (Version 6.0c, GraphPad Software, La Jolla, CA) or SPSS (Version 20.0.0, IBM Corp, Armonk, NY, USA) software. All reported P values are two-sided, and p values <.05 were considered significant.
Ethical considerations
The original study was approved by the Regional Research Ethics Committee in Lund (no 195/2005) which allowed for consent for the retro- and prospective studies at the NEP to be obtained by an opt-out arrangement, through advertisements in two daily local papers and multiple posters at the NEP itself. The study in Stockholm was approved by the Regional Research Ethics Committee (2013/495-31/3 and 2015/1374-32).
Results
Baseline characteristics
The baseline characteristics of the participants are summarized in . The gender distribution was skewed towards fewer women, similar across HCV genotypes. Interestingly, in the Stockholm cohort re-infected patients had a higher clearance rate of 67% as compared to 26% among the participants who were anti-HCV negative at enrollment. Aside from this, the clearance rates were similar across genotypes and cohorts. Among anti-HCV negative patients at enrollment developing an acute HCV infection during the study, 45% lacked IFN-λ4 (IFNL4 TTrs368234815 homozygotic), whereas the corresponding proportion among re-infected patients was 67%.
Table 1. Baseline characteristics for different cohorts and accordning to hepatitis C virus genotype.
IFNL4 and ITPase activity and spontaneous resolution of HCV infection
In anti-HCV seronegative patients at enrollment, 41 of 94 (44%) of participants lacked IFN-λ4 (IFNL4 TTrs368234815 homozygotes) vs. 22/113 (20%) of those with at least one functional gene (IFNL4 ΔGrs368234815 carriers) spontaneously cleared their infection (p < .001, Odds ratio (OR) 3.2, 95% confidence interval (CI) 1.7 to 6.0)) [].
Figure 2. (A) Percentage of spontaneous clearance of HCV infection in relation to absence of IFN-λ4 (IFNL4rs368234815 TT/TT) or presence of IFN-λ4 (IFNL4rs368234815 ΔG) for all HCV genotypes (gt), genotype 1, genotype 2/3 together and Re-infected PWID participants; statistics using Fischer’s exact test. (B) Proportion of patients with absence of IFN-λ4 (IFNL4rs368234815 TT/TT) or presence of IFN-λ4 (IFNL4rs368234815 ΔG) in four different groups; anti-HCV seronegative participants at enrollment without spontaneous clearance (SC-), Re-infected participants unable to clear their re-infection (SC + SC-), anti-HCV seronegative participants at enrollment clearing their first documented HCV infection (SC+), and Re-infected participants who had cleared at least two documented HCV infections (SC + SC+), statistics using Chi2 test.
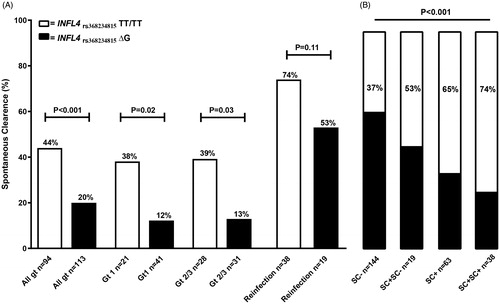
The association between absence of IFN-λ4 and greater likelihood of spontaneous resolution of HCV infection remained significant both for HCV genotype 1 infected persons (38% vs. 12% for IFNL4 TTrs1368234815 homozygotes and ΔGrs368234815 carriers respectively, p = .02; OR 4.4 (95% CI 1.2 to 16.0)), and for HCV genotype 2 or 3 when analyzed together (39% vs. 13% for IFNL4 TTrs1368234815 homozygotes and ΔGrs368234815 carriers respectively, p = .03; OR 4.4 (95% CI 1.1 to 13.8)) []. A similar, non-significant trend was seen when this latter group was further subdivided by genotypes 2 and 3; 3/6 (50%) vs. 0/5 (0%) for genotype 2 (p = .18, OR ∞ (95% CI 0.8; ∞)), and 8/22 (36%) vs. 4/26 for genotype 3 (15.0%) (p = .11, OR 3.1 (95% CI 0.8–10.5)).
Re-infected subjects had an overall higher clearance rate of 67%, and among these participants there was a similar, non-significant trend towards higher clearance rates among individuals lacking IFN-λ4 (74% vs. 53% for IFNL4 TTrs368234815 homozygotes and ΔGrs1368234815 carriers respectively, p = .1, OR 2.5 (95% CI 0.7–7.3)). Moreover, a significant increase in proportion of patients lacking IFN-λ4 (IFNL4 TTrs368234815 homozygotes) was observed when comparing the following groups: (i) anti-HCV seronegative patients at enrollment without spontaneous clearance after exposure to HCV during the follow-up study period, (ii) re-infected patients unable to clear re-infection, (iii) anti-HCV seronegative patients at enrollment clearing their first documented HCV infection, and (iv) Re-infected patients who had cleared at least two documented HCV infections [ < .001, Chi-2 test].
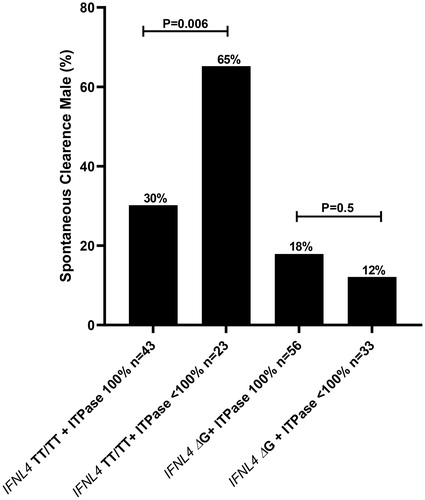
In contrast to IFNL4 genetic variants, spontaneous clearance of HCV was not significantly associated with predicted ITPase activity when analyzed independently. However, when restricting the analysis to subjects lacking IFN-λ4 (IFNL4 TTrs368234815 homozygotic), men with reduced predicted ITPase activity (defined as <100%) were significantly more likely to spontaneously resolve the HCV infection as compared to those with normal ITPase enzymatic activity (; 65% vs. 30% for ITPase <100% and 100% respectively, p = .006). This difference was not observed among men able to produce IFN-λ4 (IFNL4 ΔGrs368234815 carriers) (), nor among female patients, although it is important to note that only 51 women were enrolled in the PWID cohorts, making female-gender related subgroup analyses challenging.
Multivariate analysis and spontaneous resolution of HCV-infection
To further analyze the impact host genetic variants on spontaneous clearance, a multivariate analysis was performed including gender, age at seroconversion for HCV, ITPase activity (<100% or 100%), HCV genotype, and absence or presence of IFN-λ4 (IFNL4 TTrs1368234815 homozygote or ΔGrs368234815 carriage). In this analysis only absence of IFN-λ4 (IFNL4 TTrs368234815 homozygosity) (p = .002, adjusted OR 4.7 (95% CI 1.7–12.5)) and female gender (p = .007, adjusted OR 4.1 (95% CI 1.5–11.8)) were significantly associated with spontaneous clearance of acute incident HCV infection.
A separate multivariate analysis was performed for the whole study population, including previous anti-HCV status: (1) subjects seroconverting against HCV and (2) re-infected patients, but not including HCV genotype as this was only analyzed in the Malmö cohort. In this analysis, only absence of IFN-λ4 (IFNL4 TTrs368234815 homozygosity) (p = .0002, adjusted OR 2.9 (95% CI 1.7–5.0)) and PWID cohort (p = .0002, adjusted OR 4.0 (95% CI 1.9–8.3)) were significantly associated with spontaneous clearance of acute incident HCV infection.
Discussion
The main findings of this study were that absence of IFN-λ4 is associated with enhanced spontaneous clearance of HCV infection across genotypes 1-3, and that among men lacking IFN-λ4, reduced ITPase activity augmented the likelihood of spontaneous resolution.
IFNL4 polymorphisms have been extensively evaluated in HCV infection, especially as prognostic markers of responsiveness to interferon and ribavirin combination treatment, but also in the context of resolution of acute HCV infection [Citation2,Citation15]. Most prior studies have been conducted in geographical regions with a predominance of HCV genotype 1, but a recent meta-analysis confirmed the importance of these polymorphisms in interferon-based treatment for genotype 2 or 3 infection [Citation16]. Studies on the impact of IFNL4 polymorphisms in acute infection with HCV genotype 2 or 3 are rare, which may be explained by difficulties in identifying subjects early during infection to allow for detection of HCV RNA and genotyping. The association of absence of IFN-λ4 (CCrs12979860 carriage) and spontaneous clearance among non-1 HCV genotypes has previously been reported in a study where the majority of enrolled patients had genotype 2 or 3 infection [Citation17], and also in a cohort of patients with thalassemia and a predominance of acute HCV genotype 3 infection [Citation17,Citation18]. In the present study, PWID were longitudinally monitored at intervals of approximately 3 months leading to high precision of estimating the onset of HCV infection and also to a high HCV-RNA detection rate allowing for HCV genotyping in 121 of 139 participants from the Malmö cohort. The association between inability to produce IFN-λ4 (TTrs368234815 homozygosity) and increased likelihood of spontaneous resolution following incident exposure thus could be confirmed for HCV genotypes 1 and 2/3.
The impact of IFNL4 on spontaneous resolution in subjects who previously had cured at least one documented HCV infection was also analyzed, and an accumulation of participants lacking IFN-λ4 (TTrs368234815 homozygosity) was observed in the re-infected group, with the proportion lacking IFN-λ4 increasing among subjects that were able to also clear the reinfection. The clearance rate following reinfection among PWID lacking IFN-λ4 was relatively high at 74% as compared with 53% if one or two functional IFNL4 genes were present. These observations are in line with a well-characterized, albeit small multicenter study, where a borderline significant association with higher clearance among patients without IFN-λ4 (CCrs12979860) was reported among 28 HCV re-infected subjects [Citation19]. The difference in the present study also demonstrated borderline significance, but a clear accumulation of subjects lacking IFN-λ4 in spontaneous clearers and in individuals resolving at least two documented HCV infections was noted. The combined results of the abovementioned [Citation19] and the present study, provide increasing evidence of absence of IFN-λ4 entailing enhanced likelihood of spontaneous HCV clearance both following primary acute infection and repeated re-exposure to HCV. This may have implications regarding the need for follow-up after HCV therapy among PWID, where individuals capable of producing IFN-λ4 likely require more stringent monitoring as they are more prone to become reinfected following renewed HCV exposure.
Prior to initiating the present study, we hypothesized that among patients lacking IFN-λ4, reduced ITPase activity might be additionally beneficial based on prior studies [Citation8,Citation9]. Indeed, a highly significant association to this effect was noted among male patients lacking IFN-λ4 (TTrs368234815 homozygous), where reduced ITPase activity improved the likelihood to spontaneously resolve to 65%, as compared to 30% in those with full enzymatic activity. Interestingly, the abovementioned association was absent among female participants, which might be secondary to the small number of female participants in the present study, but might additionally by explained by well-documented gender differences in immune responses observed in numerous vaccine trials [Citation20], as well as outcome following several other viral infections, as recently demonstrated by the higher mortality noted among male COVID-19 patients [Citation21].
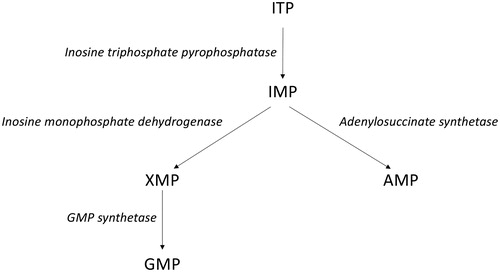
ITPase is an enzyme that metabolizes non-canonical triphosphate nucleotides (e.g., XTP and ITP, ), which otherwise might be incorporated in host or viral genomes. Additionally, ribavirin triphosphate (RTP) is dephosphorylated by ITPase in vitro to ribavirin monophosphate (RMP) at a comparable rate to ITP [Citation9]. Inosine containing single-stranded RNA is highly immunogenic and could theoretically cause a better immune response [Citation22,Citation23]. In line with this notion is the previously reported ability of ribavirin, which inhibits the enzyme inosine-5′-monophosphate dehydrogenase (IMPDH) which converts IMP to XMP, to act as an immunomodulatory compound affecting the Th1/Th2 balance [Citation24,Citation25].
Figure 4. Inosine triphosphate (ITP) to guanosine monophosphate (GMP) and adenosine monophosphate (AMP) pathways.
There are certain limitations in the present study. Patients were defined as spontaneous clearers based on undetectable HCV RNA in one sample. The samples were obtained approximately one year after seroconversion in the Malmö cohort and >5 months past reinfection, and thus the two cohorts had slightly differing sampling timepoints. Also, during the first year we have earlier reported very divergent patterns of viremia with some patients still having very low-level HCV viremia at their one-year visit [Citation26]. The sera also were diluted 1/10 prior to HCV RNA analysis in the Malmö cohort because of limited sample volume availability, which potentially may have led to a slightly overestimated clearance rate. Additionally, genotyping was performed only once in the Malmö cohort and was not assessed in the Stockholm cohort. As the incidence of re-infection among these PWID was 31/100 person-years with a potential risk of misclassifying a spontaneous clearer with re-infection with a new genotype as being a chronically infected person. In 18 individuals, the HCV genotype could not be determined, and 9 of these participants never had detectable HCV-RNA in spite of documented seroconversion, presumably due to a missed short viremia peak occurring between available samples.
In conclusion, the present study demonstrated that IFNL4 genetic variants resulting in inability to produce IFN-λ4 are linked with improved likelihood of spontaneous clearance both following primary acute infection and repeated re-exposure to HCV across genotypes 1–3. Additionally, among men lacking IFN-λ4, reduced ITPase activity significantly augmented the likelihood of resolution.
Acknowledgements
We thank Vilma Molnegren, Ludmila Adamek, and Anette Roth for technical assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alberti A, Chemello L, Benvegnu L. Natural history of hepatitis C. J Hepatol. 1999;31:17–24.
- Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801.
- Shebl FM, Pfeiffer RM, Buckett D, et al. IL28B rs12979860 genotype and spontaneous clearance of hepatitis C virus in a multi-ethnic cohort of injection drug users: evidence for a supra-additive association. J Infect Dis. 2011;204(12):1843–1847.
- Prokunina-Olsson L, Muchmore B, Tang W, et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet. 2013;45(2):164–171.
- Fellay J, Thompson AJ, Ge D, et al. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464(7287):405–408.
- Sumi S, Marinaki AM, Arenas M, et al. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111(4–5):360–367.
- Shipkova M, Lorenz K, Oellerich M, et al. Measurement of erythrocyte inosine triphosphate pyrophosphohydrolase (ITPA) activity by HPLC and correlation of ITPA genotype-phenotype in a Caucasian population. Clin Chem. 2006;52(2):240–247.
- Rembeck K, Waldenstrom J, Hellstrand K, et al. Variants of the inosine triphosphate pyrophosphatase gene are associated with reduced relapse risk following treatment for HCV genotype 2/3. Hepatology. 2014;59(6):2131–2139.
- Nystrom K, Wanrooij PH, Waldenstrom J, et al. Inosine triphosphate pyrophosphatase dephosphorylates ribavirin triphosphate and reduced enzymatic activity potentiates mutagenesis in hepatitis C virus. J Virol. 2018;92(19):e01087-18.
- Maheshwari A, Ray S, Thuluvath PJ. Acute hepatitis C. Lancet. 2008;372(9635):321–332.
- Blome MA, Bjorkman P, Flamholc L, et al. Minimal transmission of HIV despite persistently high transmission of hepatitis C virus in a Swedish needle exchange program. J Viral Hepat. 2011;18(12):831–839.
- Kaberg M, Naver G, Hammarberg A, et al. Incidence and spontaneous clearance of hepatitis C virus (HCV) in people who inject drugs at the Stockholm Needle Exchange – importance for HCV elimination. J Viral Hepat. 2018;25(12):1452–1461.
- Abdel-Hamid M, El-Daly M, Molnegren V, et al. Genetic diversity in hepatitis C virus in Egypt and possible association with hepatocellular carcinoma. J Gen Virol. 2007;88(5):1526–1531.
- Rembeck K, Alsio A, Christensen PB, et al. Impact of IL28B-related single nucleotide polymorphisms on liver histopathology in chronic hepatitis C genotype 2 and 3. PLoS One. 2012;7(1):e29370.
- Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401.
- Gauthiez E, Habfast-Robertson I, Rueger S, the Swiss Hepatitis C Cohort Study, et al. A systematic review and meta-analysis of HCV clearance. Liver Int. 2017;37(10):1431–1445.
- Grebely J, Page K, Sacks-Davis R, the InC3 Study Group, et al. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59(1):109–120.
- Biswas A, Firdaus R, Gupta D, et al. Interferon lambda3 gene (IL28B) is associated with spontaneous or treatment-induced viral clearance in hepatitis C virus-infected multitransfused patients with thalassemia. Transfusion. 2017;57(6):1376–1384.
- Sacks-Davis R, Grebely J, Dore GJ, et al. Hepatitis C virus reinfection and spontaneous clearance of reinfection–the InC3 study. J Infect Dis. 2015;212(9):1407–1419.
- Klein SL, Jedlicka A, Pekosz A. The Xs and Y of immune responses to viral vaccines. Lancet Infect Dis. 2010;10(5):338–349.
- Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention. [The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China]. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145–151.
- Liao JY, Thakur SA, Zalinger ZB, et al. Inosine-containing RNA is a novel innate immune recognition element and reduces RSV infection [Research Support, N.I.H., Intramural]. PloS One. 2011;6(10):e26463.
- Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–457.
- Brenndorfer ED, Brass A, Karthe J, et al. Cleavage of the T cell protein tyrosine phosphatase by the hepatitis C virus nonstructural 3/4A protease induces a Th1 to Th2 shift reversible by ribavirin therapy. JI. 2014;192(4):1671–1680.
- Hultgren C, Milich DR, Weiland O, et al. The antiviral compound ribavirin modulates the T helper (Th) 1/Th2 subset balance in hepatitis B and C virus-specific immune responses. J Gen Virol. 1998;79(10):2381–2391.
- Alanko Blome M, Bjorkman P, Molnegren V, et al. Hepatitis C viremia patterns in incident hepatitis C infection and one year later in 150 prospectively tested persons who inject drugs. PLoS One. 2014;9(5):e97022.