Abstract
Objective
Assessment of the anatomical colorectal segment of polyps during colonoscopy is important for treatment and follow-up strategies, but is largely operator dependent. This feasibility study aimed to assess whether, using images of a magnetic endoscope imaging (MEI) positioning device, a deep learning approach can be useful to objectively divide the colorectum into anatomical segments.
Methods
Models based on the VGG-16 based convolutional neural network architecture were developed to classify the colorectum into anatomical segments. These models were pre-trained on ImageNet data and further trained using prospectively collected data of the POLAR study in which endoscopists were using MEI (3930 still images and 90,151 video frames). Five-fold cross validation with multiple runs was used to evaluate the overall diagnostic accuracies of the models for colorectal segment classification (divided into a 5-class and 2-class colorectal segment division). The colorectal segment assignment by endoscopists was used as the reference standard.
Results
For the 5-class colorectal segment division, the best performing model correctly classified the colorectal segment in 753 of the 1196 polyps, corresponding to an overall accuracy of 63%, sensitivity of 63%, specificity of 89% and kappa of 0.47. For the 2-class colorectal segment division, 1112 of the 1196 polyps were correctly classified, corresponding to an accuracy of 93%, sensitivity of 93%, specificity of 90% and kappa of 0.82.
Conclusion
The diagnostic performance of a deep learning approach for colorectal segment classification based on images of a MEI device is yet suboptimal (clinicaltrials.gov: NCT03822390).
Introduction
The role of accurate polyp and colorectal cancer (CRC) anatomical localization during colonoscopy has become increasingly important in the modern endoscopy era [Citation1]. Determining the precise location of colorectal polyps and cancers is crucial for selecting the optimal technique for endoscopic resection, guiding subsequent surgical resection, determining the optimal surveillance interval, and for relocating the polypectomy scar during follow-up colonoscopy. However, assessment of the anatomical location during colonoscopy is subject to inter-observer variation, and affected by expertise and mood of the endoscopist [Citation2–12]. Inaccurate determination of the location of polyps may be avoided in many cases by endoluminal tattooing of lesions throughout endoscopy. However, small polyps containing unsuspected cancer will usually not be tattooed and besides, tattooing is regularly forgotten. This is particularly the case when multiple polyps are resected, which is common in bowel cancer screening patients. Objectively and automatically determining the anatomical location (i.e., colorectal segment) during colonoscopies could optimize treatment and surveillance strategies.
A possible way to reduce the inter-observer variation of colorectal segment classification may be to use artificial intelligence (AI) techniques, given its potential to recognize informative patterns in imagery. The recent success of deep learning for various applications has greatly leveraged this potential [Citation13,Citation14]. To date however, little AI-related research has been done on the topic of colorectal segment classification [Citation15–17]. Furthermore, to enhance the precise classification of colorectal segments, images of a magnetic endoscope imaging (MEI) positioning device (e.g. ScopeGuide®, Olympus, Corporation), may be usefull [Citation3]. A MEI positioning device is an accessory tool that provides on a monitor images of the position and configuration of the endoscope inside the colon. However, when using a MEI positioning device, the interpretation of the anatomical location is still done by the endoscopist. In this study, we therefore aimed to develop an accurate, objective and automatic method to classify the colorectum into anatomical segments using deep learning and images of the MEI positioning device.
Methods
Setting and study design
This study was designed to evaluate the diagnostic accuracy of deep learning models for the classification of the colorectal segment, based on images of the MEI positioning device. The study is reported according to the STARD statements [Citation18]. For this study we used the data of a prospective multicenter study in which a computer-aided diagnosis (CADx) for optical diagnosis of diminutive polyps was developed, validated and benchmarked against the performance of screening endoscopists (POLyp Artificial Recognition [POLAR] system) (Houwen B, 2022, unpubl. data). This study was conducted from October 2018 to September 2021 in eight regional Dutch hospitals and one academic Spanish hospital in partnership with ZiuZ Visual Intelligence (Gorredijk, the Netherlands). Data were collected during colonoscopies in the context of the bowel cancer screening and surveillance program or for the evaluations of symptoms. The detailed methods and results of this study are described elsewhere (online). For this study, we used prospectively collected training data from one center in which the MEI device was used during the study procedures.
Outcome measures and definitions
The main outcome of this study was the accuracy of the models for predicting the correct anatomical segment of the colorectum. Accuracy was defined as the percentage of correctly predicted colorectal segments of the model compared to the reference standard. The colorectal segment assignment by the endoscopist during colonoscopy was used as reference standard. Secondary outcomes such as specificity, sensitivity, F1-score, the weighted kappa score and in particular the confusion matrix were used to provide more insight not only about the broad performance of the model, but also on the category it could be confused with. This is important due to the category imbalance within data. The diagnostic accuracy tests were performed for a 5-class colorectal segment division (cecum, ascending colon/hepatic flexure, the transverse colon, the colon descendens/splenic flexure and the rectum/sigmoid) and 2-class colorectal segment division (proximal to rectosigmoid and rectosigmoid).
Data collection procedure and data preparation
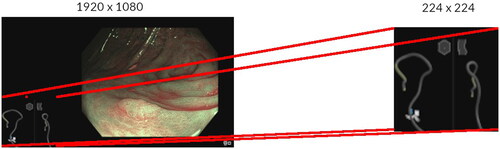
Data in the POLAR study were prospectively collected following a standardized image acquisition protocol. The endoscopist had to take an image of each detected lesion from the front with high-definition white-light endoscopy (HD-WLE) as well as with narrow-band imaging (NBI) without magnification (Olympus CF-HQ190L colonoscope with a resolution of 1920 × 1080 pixels). When an image was taken by the endoscopist, 22 video frames were as well recorded directly before and directly after the image (in a period of 1s). The images and video frames, together referred as snapshots, were saved to a storage system used for this study. A snapshot contained the output of the colonoscope camera and the output of MEI device. Before training the models, all individual snapshots were cropped to only the pixels containing the MEI device output, and resized to 224 × 224 pixels (). Endoscopists of the POLAR study that were performing the colonoscopy annotated the colorectal segment of each polyp directly during the procedure. Endoscopists could choose between cecum, ascending colon, hepatic flexure, transverse colon, splenic flexure, descending colon, sigmoid, rectosigmoid and rectum. In order to properly evaluate the models for colorectal segment classification, snapshots that did not contain both a lateral- and anteroposterior view from the MEI device were excluded as were snapshots in which the MEI device had failed to produce an output due to technical issues (Supplementary Figure 1). Snapshots in which the endoscopist annotated a polyp with two colorectal segments were also left out.
Figure 1. Examples of a snapshot (All individual snapshots were cropped to the pixels containing the MEI positioning device output).
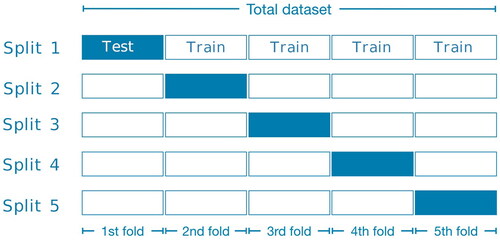
After performing data-augmentation by vertical- and horizontal shift, the dataset was split into five groups for five-fold cross validation (). In order not to introduce bias, snapshots from the same procedure could not be in both the train-folds and test-fold. At each iteration, all snapshots from one of these folds were being used as the test set and the snapshots from the other four folds were used to train the model. After five iterations all snapshots in the dataset have been used exactly once for the evaluation of the model and we could compute evaluation metrics across the whole dataset. To increase the robustness of the analyses, the five-fold cross validation was repeated five times.
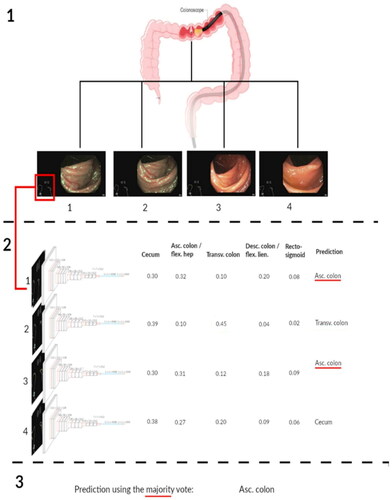
Model development
Supplementary text 1 provides an extensive description of the model development. An off-the-shelf VGG-16 CNN architecture was used as the core of our model, i.e., a model to classify the colon into anatomical segments. The VGG-16 we used was pre-trained on the ImageNet dataset, a dataset with more than 14 million hand-annotated images from a broad range of object classes [Citation19,Citation20]. Subsequently, these models were further trained using prospectively collected data of the POLAR study in which endoscopists were using the MEI device. For each MEI device snapshot, the model produced a probability score between 0 and 1, representing the probability that this snapshot came from a particular colorectal segment category. This sample-based model used the highest probability score to determine the colorectal segment category (i.e., for the 5-class division at least one l segment had a prediction score of ≥0.20). Subsequently, several post-processing strategies were assessed to improve the model’s performance. The majority vote strategy was used as final polyp-based model, since this model slightly outperformed the other models in the 5-fold cross validation setting () [Citation21]. By adopting the majority vote strategy, the colorectal segment category snapshot predictions per polyp were added up together. The most dominant snapshot prediction per polyp, was used for the definitive colorectal segment category (i.e., polyp-based model). The model was developed in Python using Keras with the TensorFlow 2 backend and ran on the Dutch national compute cluster LISA [Citation22].
Statistical analysis
Diagnostic test accuracies (accuracy, specificity, sensitivity, F1-score and weighted kappa) for colorectal segment classification for the sample-based and polyp-based model were calculated. The colorectal segment assignment by the endoscopists was used as the reference standard. Answers were dichotomized to a positive outcome for each colorectal segment category of interest and a negative outcome for the other colorectal segment categories. The average results over five runs of the five-fold cross validation were used for the diagnostic accuracy calculations. We decided to include the Quadratic weighted Cohen’s Kappa score (QW-Kappa score) which takes the sorted nature of categories in an ordinal classification into account. Analysis was performed in statistical software R (version 4.0.3).
Results
Baseline characteristics
The initial POLAR dataset comprised 115,412 MEI snapshots consisting of both images and video frames. After the initial data set cleaning, 94,081 snapshots remained that could be included for the training and testing of our colorectal segment classification models (3930 images and 90,151 corresponding video frames). These snapshots were collected during 416 different colonoscopies with 1196 polyps. The mean number of snapshots per polyp in the data set was 89 (Q1–Q3 range 52–124) (Supplementary Figure 2). presents the distribution of polyps and snapshots in the different colorectal segments.
Table 1. Distribution of the included data.
Diagnostic accuracy of the snapshot-based and polyp-based model for colorectal segment classification
illustrates the colorectal segment classification performance for the snapshot-based model (i.e., non-polyp based) and the polyp-based model. For both the 5-class and 2-class segment division, the polyp-based model performed slightly better than the snapshot-based model. For the 5-class colorectal segment division, the sample-based model classified the colorectal segment in 57,389 of the 94,081 snapshots correctly, corresponding to an overall accuracy of 61%, sensitivity of 61% and specificity of 89%. For the 2-class colorectal segment division, the model classified the colorectal segment in 86,554 of the 94,081 snapshots correctly, corresponding to an overall accuracy of 92%, sensitivity of 92% and specificity of 88%. When the colon was divided into five segments, the polyp-based model correctly predicted the colon segment category in 753 of the 1196 polyps, corresponding to an overall accuracy of 63%, sensitivity of 63% and specificity of 89%. For the 2-class colorectal segment division, the polyp-based model classified the colorectal segment in 1112 of the 1196 polyps correctly, corresponding to an accuracy of 93%, sensitivity of 93% and specificity of 90%.
Table 2. Classification performance for snapshot-based strategy and polyp-based strategy, mean (SD)a.
Diagnostic accuracy of polyp-based model for classification per colorectal segment
shows the classification performance of the polyp-based model per colorectal segment. The model achieved in the highest performance in the most prevalent colorectal segment classes (i.e., rectosigmoid and ascending colon). When the colon was divided into five colorectal segments, the model achieved in the colorectal segment classes rectosigmoid and ascending colon a sensitivity of at least 69% and a specificity of at least 80%. In the cecum, transverse and descending colon the model achieved only a sensitivity between 38% and 46% and a specificity between 90% and 96%. shows the confusion matrix of polyp’s colon segment classification for the 5-class division. Colorectal segments anatomically closer to each other were more often interchanged.
Figure 4. Confusion matrix between the classified colorectal segment and annotated colorectal segment.
For the 5-class and 2-class colorectal segment classification confusion matrix, the polyp-based majority vote strategy outcomes from one of the five runs was used.
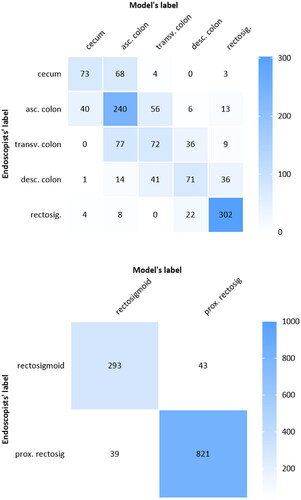
Table 3. Classification performance per colonic segmenta.
Discussion
In the present study, we propose a novel and objective method for classification of colorectal segments during colonoscopy using deep learning based on images of the magnetic endoscope imaging (MEI) positioning device. We constructed convolutional neural network (CNN) models based on 3930 still images and 90,151 corresponding video frames and showed its diagnostic performance. When the colon was divided into five anatomical segments, the best performing model achieved an accuracy of 63%, sensitivity of 63% and a specificity of 89%. With a simplified but still clinical useful division of the colon in two colorectal segments (rectosigmoid versus the rest), the performance of the model improved to an accuracy of 93%, sensitivity of 93% and a specificity of 90%, which may be clinically useful.
The diagnostic performance of the model differed according to the different colorectal segment categories. For a five-class division, the colorectal segment categories with the highest number of images (i.e., rectosigmoid and ascending colon) had sensitivities above 69%, whereas it had lowest specificity at 80%. By contrast, the colorectal segment categories with the lowest number of images (the cecum, transverse colon, descending colon) had high specificities but sensitivities below 46%. The relatively low sensitivities in the colorectal segments with the lowest number of polyps (i.e., minority classes) are most probably due to overfitting on the colorectal segments with the highest number of polyps (i.e., the majority classes). Multiple strategies for preventing the overfitting on the majority classes can be studied, from under- or oversampling the imbalanced classes, to using a weighted loss function. Although implementing these techniques would improve the class wise accuracy, the overall accuracy would probably not be affected. Since all colorectal segment classes are equally important, class-balancing techniques are not desirable for this classification problem.
To enhance the accuracy for automated colorectal segment classification, recurrent neural networks (i.e., Long Short-Term Memory networks) may be useful. Recurrent neural networks operate on sequences of multiple images and can learn features from the patterns within a whole sequence. Since we had a median of 89 snapshots per polyp, learning such patterns could be beneficial. Our algorithm utilizes the information that multiple, subsequent images refer to the same polyp only after the predictions for single images are made and only in order to aggregate results. As such, it will not be able to learn any sequence-wide patterns. The PlaNet CNN developed by Google engineers for geolocating photos from the outside world achieved good results using this method [Citation21]. Another promising opportunity to improve the accuracy using deep learning might be by the use of temporal information- knowing which colorectal segment was visited before [Citation23]. This temporal information plays a big role in the endoscopists’ ability to predict its current position, and might also be used in a deep learning approach for this problem. Another future research direction could lay more into combining colonoscopy and/or MEI with other modalities that show 3-dimensional information may be useful. Liu et al. and more recently Yao et al. used the optical flow (i.e., motion) between sequential video frames to track the location on the tip of the colonoscope [Citation15,Citation16]. Using the camera trajectory they constructed a 3-dimensional map of the visualized colon during each colonoscopy, a map in which polyps were located. With manually annotated frames, they achieved in their study a 75% accuracy for classification with a 6-class colon segment division. These results seem, just like ours, promising but also show that classifying the correct colorectal segment category is not an easy task to solve.
The strength of this study is primarily its novelty. To our knowledge developing an objective and automatic method to classify the colorectal segment, combining deep learning techniques and snapshots of a MEI positioning device, has not been done before. If this method with MEI proofs to be accurate, endoscopist could objectively and easily classify the anatomical location of polyps with during colonoscopy, thereby improving the clinical management of polyps. Another future application of colorectal segment classification would be to automatically generate endoscopy reports and thereby reducing the workload for endoscopists. We recognize there are also several limitations to consider regarding our study. The most important one is that our reference standard, the colorectal segment prediction by a single endoscopist, may not itself have been accurate, given previous reports of 11–21% inaccuracy when compared with surgical classificaiton [Citation9,Citation24]. Additionally, the reference standard itself might be biased, since the endoscopist could also have used the MEI device during endoscopy to determine the colorectal segment. Using the colorectal segment as determined in the surgical resection of polyps as ground truth would improve the reliability of our models. However, it will be difficult to collect a large amount of data on surgically resected of polyps as ground truth. Another limitation is our relatively small dataset size obtained at a single institution (so lack of an external test set), in which the distribution of each colorectal segments was unbalanced, which resulted in overfitting on the majority classes. As shown in Supplementary Figure 3, the MEI device produced sometimes very complex snapshots, and these snapshots were usually predicted incorrectly by our models. We kept them in our training- and test data as we were unable to prove these complex snapshots were technical failures of the MEI device. Since too few of these complex MEI device snapshots were present in the training set to effectively train on, they presumably affected the accuracy of our models significantly. Having more data available would probably improve the robustness and performance and possibly even diminish the negative effects of those outliers on the performance.
To conclude, classifying the anatomical colorectal segment during colonoscopy is greatly operator dependent. The present study revealed the diagnostic performance of a CNN model for colorectal segment classification based on images of a MEI device. This could be the first step towards an automatic and objective alternative for the classification of colorectal segments during colonoscopy. More research and development is needed before such a model could be implemented in endoscopy systems and put into production. More data from multiple centers, and using temporal information or more sophisticated networks like LSTM should be researched with a more robust reference standard.
Author contributions
Conception and design: BH, FH, IG, YH, ED; Data acquisition: all authors; Data analysis and interpretation: all authors; Drafting the manuscript: BH, FH; Critical revision of the manuscript: all authors; Supervision: IG, YH, PF, ED.
| Abbreviations | ||
| AI | = | Artificial intelligence |
| CRC | = | Colorectal cancer |
| CI | = | confidence interval |
| HD | = | high-definition |
| CNN | = | convolutional neural network |
Supplemental Material
Download PDF (409.5 KB)Disclosure statement
ED has received speaker’s fees from Roche (2018), Norgine (2019), Olympus and GI Supply (both 2019–2020), and Fujifilm (2020), and has provided consultancy to Fujifilm (2018), CPP-FAP (2019), GI Supply (2019–2020), Olympus (2020 to present), PAION and Ambu (both 2021); she received a research grant from Fujifilm (2017–2020) and her department has equipment on loan from Fujifilm (2017 to present) and Olympus (2021). PF received research support from Boston Scientific and a consulting fee from Olympus and Cook Endoscopy. MP has received speaker’s fees from Casen Recordati (2016–2019), Olympus (2018), Jansen (2018), Norgine (2016–2021) and Fujifilm (2021), an editorial fee from Thieme as an Endoscopy co-Editor (2015–2021) and has provided consultancy to Norgine Iberia (2015–2019) and GI Supply (2019); she received a research grant from Fujifilm (2019–2021), Casen recordati (2020) and ZiuZ (2021) and her department has equipment on loan from Fujifilm (2017–present). The other authors have no relevant disclosures to report.
Additional information
Funding
References
- Hassan C, Quintero E, Dumonceau JM, et al. Post-polypectomy colonoscopy surveillance: European Society of Gastrointestinal Endoscopy (ESGE) guideline. Endoscopy. 2013;45(10):842–851.
- Borda F, Jiménez FJ, Borda A, et al. Endoscopic localization of colorectal cancer: study of its accuracy and possible error factors. Rev Esp Enferm Dig. 2012;104(10):512–517.
- Ellul P, Fogden E, Simpson C, et al. Colonic tumour localization using an endoscope positioning device. Eur J Gastroenterol Hepatol. 2011;23(6):488–491.
- Feuerlein S, Grimm LJ, Davenport MS, et al. Can the localization of primary colonic tumors be improved by staging CT without specific bowel preparation compared to optical colonoscopy? Eur J Radiol. 2012;81(10):2538–2542.
- Louis MA, Nandipati K, Astorga R, et al. Correlation between preoperative endoscopic and intraoperative findings in localizing colorectal lesions. World J Surg. 2010;34(7):1587–1591.
- Moug SJ, Fountas S, Johnstone MS, et al. Analysis of lesion localisation at colonoscopy: outcomes from a multi-Centre U.K. study. Surg Endosc. 2017;31(7):2959–2967.
- Neri E, Turini F, Cerri F, et al. Comparison of CT colonography vs. conventional colonoscopy in mapping the segmental location of Colon cancer before surgery. Abdom Imaging. 2010;35(5):589–595.
- O'Connor SA, Hewett DG, Watson MO, et al. Accuracy of polyp localization at colonoscopy. Endosc Int Open. 2016;4(6):E642–46.
- Piscatelli N, Hyman N, Osler T. Localizing colorectal cancer by colonoscopy. Arch Surg. 2005;140(10):932–935.
- Solon JG, Al‐Azawi D, Hill A, et al. Colonoscopy and computerized tomography scan are not sufficient to localize right‐sided colonic lesions accurately. Colorectal Dis. 2010;12(10 Online):e267-72–e272.
- Stanciu C, Trifan A, Khder SA. Accuracy of colonoscopy in localizing colonic cancer. Rev Med Chir Soc Med Nat Iasi. 2007;111(1):39–43.
- Vaziri K, Choxi SC, Orkin BA. Accuracy of colonoscopic localization. Surg Endosc. 2010;24(10):2502–2505.
- Misawa M, Kudo SE, Mori Y, et al. Current status and future perspective on artificial intelligence for lower endoscopy. Dig Endosc. 2021;33(2):273–284.
- Ahmad OF, Soares AS, Mazomenos E, et al. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol. 2019;4(1):71–80.
- Liu J, Subramanian KR, Yoo TS. An optical flow approach to tracking colonoscopy video. Comput Med Imaging Graph. 2013;37(3):207–223.
- Yao H, Stidham RW, Gao Z, et al. Motion-based camera localization system in colonoscopy videos. Med Image Anal. 2021;73(4):102180.
- Saito H, Tanimoto T, Ozawa T, et al. Automatic anatomical classification of colonoscopic images using deep convolutional neural networks. Gastroenterol Rep (Oxf). 2021;9(3):226–233.
- Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527.
- Russakovsky O, Deng J, Su H, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252.
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Comput. Vis. Pattern Recognit. 2014. arXiv:1409.1556.
- Weyand T, Kostrikov I, Philbin J. Planet-photo geolocation with convolutional neural networks. In: European Conference on Computer Vision. Los Angeles: Springer; 2016.
- LISA. A Beowulf cluster computer consisting of several hundreds of multi-core nodes running the Debian Linux operating system [Internet] [cited 2022 June 14]. Available from: https://servicedesk.surfsara.nl/wiki/display/WIKI/Lisa.
- Bodenstedt S, Wagner M, Katić D, et al. Unsupervised temporal context learning using convolutional neural networks for laparoscopic workflow analysis. Comput. Vis. Pattern Recognit. 2017. arXiv:1702.03684.
- Cho YB, Lee WY, Yun HR, et al. Tumor localization for laparoscopic colorectal surgery. World J Surg. 2007;31(7):1491–1495.