Abstract
Background
The etiopathogenesis of diverticular disease is unknown.
Objective
To compare the fecal and mucosa-associated microbiota between participants with and without diverticulosis and participants who later developed diverticulitis versus those that did not from a population-based study.
Methods
The PopCol study, conducted in Stockholm, Sweden, invited a random sample of 3556 adults to participate, of which 745 underwent colonoscopy. Overall, 130 participants (17.5%) had diverticulosis. 16S rRNA gene sequencing was conducted on available sigmoid biopsy samples from 529 and fecal samples from 251 individuals. We identified individuals who subsequently developed acute diverticulitis up to 13 years after sample collection. In a case-control design matching for gender, age (+/−5 years), smoking and antibiotic exposure, we compared taxonomic composition, richness and diversity of the microbiota between participants with or without diverticulosis, and between participants who later developed acute diverticulitis versus those who did not.
Results
No differences in microbiota richness or diversity were observed between participants with or without diverticulosis, nor for those who developed diverticulitis compared with those who did not. No bacterial taxa were significantly different between participants with diverticulosis compared with those without diverticulosis. Individuals who later developed acute diverticulitis (2.8%) had a higher abundance of genus Comamonas than those who did not (p = .027).
Conclusions
In a population-based cohort study the only significant difference was that those who later develop diverticulitis had more abundance of genus Comamonas. The significance of Comamonas is unclear, suggesting a limited role for the gut microbiota in the etiopathogenesis of diverticular disease.
Introduction
Diverticular disease (DD) is a common and costly disease with an estimated economic burden of four billion USD per year in the USA [Citation1]. Diverticula form when the colonic mucosa extrudes between the muscular layers at the vascular entry site [Citation2]. Most individuals are asymptomatic but some develop complications in the form of diverticular hemorrhage, acute diverticulitis, peritonitis and perforation [Citation2]. Symptoms of abdominal pain and change in bowel habits in an individual with diverticulosis is often referred to as symptomatic uncomplicated diverticular disease (SUDD) [Citation3].
The etiopathogenesis of diverticular disease is largely unknown but genetics, smoking, overweight and possible differences in microbiota composition are plausible causes [Citation4]. Studies of the microbiota and DD are however few and most have a low number of participants. In most studies only fecal microbiota has been investigated, but a few studies have looked at mucosa-associated microbiota in biopsies collected during colonoscopy. Jones et al. investigated mucosal biopsies in 226 individuals with diverticulosis and 309 without diverticulosis and observed a higher abundance of phylum Proteobacteria and genus Comamonas. However, the association was very weak, and the authors concluded that the mucosal-adherent microbiota community composition is unlikely to play a role in the development of diverticulosis [Citation5]. Daniels et al. investigated fecal samples from 31 individuals with CT-confirmed asymptomatic left-sided acute diverticulitis compared with 25 controls and reported that the Shannon index indicated a higher diversity in individuals with acute diverticulitis for Proteobacteria and all phyla combined [Citation6]. Tursi et al. analyzed fecal microbiota from 15 patients with SUDD, 13 with diverticulosis and 16 healthy controls. The abundance of Akkermansia muciniphila was significantly higher in the diverticulosis group than healthy controls [Citation7]. Barbara et al. compared mucosal and fecal microbiota from 16 with diverticulosis, 14 healthy controls and 8 individuals with SUDD. Patients with diverticulosis and SUDD had a lower relative abundance of Clostridium cluster IV bacteria in their fecal samples [Citation8]. Thus, a consistent pattern of microbial changes in the conditions associated with diverticula does not emerge from the available data; rather, different studies highlight changes in bacteria from a variety of phyla. Furthermore, these studies included patients and healthy controls and as such none of the published studies on microbiota composition and diverticular disease are population-based.
We used fecal and endoscopic biopsy samples from the Swedish population-based colonoscopy study (PopCol) [Citation9] to study microbiota composition in individuals with diverticulosis compared with those without diverticulosis. Furthermore, we analyzed the association between microbiota composition and the later development of diverticulitis.
Materials and methods
This nested case-control study is a part of the Swedish population-based colonoscopy study (PopCol), which collected samples between 2002 and 2006, aiming to investigate gastrointestinal (GI) symptoms and pathology in the general population. A detailed description of participant recruitment for the study as well as its methodology has been previously described [Citation9]. The baseline study was approved by the local ethics committee at Karolinska Institutet (D.nr. 394/01). The linkage and patient file review were approved by the regional ethics board in Stockholm (D. nr. 2016/228-31/2 and 2017/603-32).
Study population
A random sample of 3556 individuals (aged 18–70 years) from Maria and Katarina parishes, two adjacent urban districts in Stockholm, Sweden was invited to participate in the study. The study has previously been described in detail [Citation9]. The study region is socio demographically similar to Sweden overall, with the mean income comparable to the mean income in Sweden although the education level was higher in the two urban districts. Of 2293 respondents, 1673 were reached by telephone, of which 1244 were scheduled for a preparation visit to a gastroenterologist where the use of all medication, smoking habits, height and weight was recorded. BMI was calculated as weight in kg divided by height in meters squared. The participants were asked specifically for recent use of antibiotics, defined as any antibiotics taken in the last 3 months. Fecal samples were collected by the participants at home between the preparation visit and the commencement of bowel cleansing prior to the colonoscopy.
A total of 745 participants with available validated symptom survey data agreed to undergo a colonoscopy with biopsies. The cecum or ileum was reached in 702 of the 745 (94%) colonoscopies. Of those that underwent colonoscopy, 37 participants had previously had an appendectomy, four had undergone cholecystectomy, one had surgery for gastrointestinal reflux disease and one had undergone ileocecal resection. Those that had the colonoscopy were older (51.7 vs 48.7 years) than those that did not undergo a colonoscopy, had more abdominal symptoms (68% vs 55%) but there was a similar sex distribution (57.2% vs 57.3% women) [Citation9]. The colonoscopies were performed by 12 senior gastroenterologist that were instructed to record all pathological findings in each segment and to record the presence of diverticula specifically. Information regarding diverticulosis status was missing for three individuals, and 130 individuals out of the remaining 742 had diverticulosis (17.4%). None had diverticulitis or a previous history of diverticulitis. All 130 individuals with diverticulosis had involvement in the sigmoid colon. Involvement in the descending colon was found in 12.7% of individuals, transverse colon in 13.6%, ascending colon in 4.0% and cecum in 3.2% [Citation10]. The prevalence of diverticulosis increased with age (1.3% before 40 years, 13.1% between 40 and 60 and 36.3% older than 60) [Citation10].
A total of 715 individuals had sigmoid biopsies taken. Fecal samples were added later to the study protocol and 352 fecal samples were collected [Citation11]. After removing samples with insufficient PCR amplification, 529 sigmoid biopsies and 251 fecal samples yielded acceptable microbiota data ().
In July 2016, study data were linked with health care data using the personal identity number assigned to all Swedish citizens. Using ICD-10 codes all out-patients and in-patients visits with a code for diverticular disease (K57) were identified. Of the 2293 individuals who took part in the PopCol study 118 individuals had at least one healthcare visit with an ICD-code for diverticular disease during the 10-year follow-up period (2006–2015). Patient files from all individuals with a code of diverticular disease were retrieved and reviewed to identify those that did develop acute diverticulitis. Among those that did a colonoscopy in the PopCol study and had data on the presence of diverticula (n = 742), 21 developed diverticulitis. Diverticulitis was diagnosed by computer tomography or acute operation in 16 cases. In the remaining five cases the diagnosis was based on clinical diagnosis.
Cases with diverticulosis were matched with individuals without diverticulosis. In addition, the cases and controls were matched by age (± 5 years), sex, current smoking (yes/no), and recent antibiotic use to obtain 2–5 controls per case.
Cases who later developed acute diverticulitis were matched to controls with diverticulosis that did not later develop acute diverticulitis. The cases and controls were matched by age (± 5 years), sex, current smoking status, recent antibiotic use and whether or not they had diverticulosis during the original colonoscopy to obtain 2–5 controls per case.
DNA extraction, 16S rRNA gene amplification and sequencing
All samples were handled and analyzed as previously described [Citation11]. In brief, biopsy samples were amplified from 170 ng of DNA for 30 cycles of PCR amplification. Fecal samples were amplified from 50 ng of DNA for 25 cycles of PCR. All sequencing was conducted on an Illumina MiSeq with 2 × 300 bp reads. Cutadapt v1.14 was used to trim 3′ bases with a Phred score lower than 15 and 5′ primer sequences [Citation12]. Reads that did not contain both primer sequences, that had less than 120 bp left after trimming or that contained more than 3 N-calls were discarded. Reads were merged and dereplicated with Usearch v10.0.240 [Citation13]. Singleton sequences and non-merging reads were discarded. ASV (amplified sequence variants) were picked with the unoise3 denoising algorithm. All merged reads from each sample were mapped back to the centroids at a minimal identity of 98% for quantification. Taxonomy was assigned to centroids by mapping them to a curated taxonomy of the SILVA v132 database as implemented in the DADA2 package (v1.6.0) with functions assign Taxonomy and addSpecies [Citation14,Citation15]. Samples were kept if they reached 12,000 approved reads. ASV were kept if their total counts reached at least 5% of the total number of samples, that is, 29 counts or higher.
Statistical analysis
ASV-level data was submitted to Maaslin2 to identify specific taxa associated with the development of disease [Citation16], either diverticulosis or diverticulitis, from either fecal or biopsy samples. ASV were kept in the analysis if they were found in at least 10% of the relevant samples with at least 1 count. Total sum normalization followed by log transformation was applied. ASV were fitted with diverticulosis or diverticulitis. The same analysis was conducted after collapsing ASV to their most complete taxonomic assignment.
Alpha (within-sample) diversity was characterized as Chao1 species-richness and as Shannon’s entropy. Differences in alpha diversity and in the relative abundance of selected clades were calculated as Welch’s t-test. Beta (across-sample) diversity was calculated as Bray–Curtis dissimilarity on ASV. Visualizations of beta diversity are based on non-metric multidimensional scaling (NMDS).
For demographic data continuous variables are presented as median and categorical variables as percentages. Differences in continuous variables were tested using the Mann–Whitney rank sum test, and a chi-square test was used to test differences in categorical variables.
The significance level for all tests is set as 0.05, with Benjamini–Hochberg adjustment for multiple testing correction. All statistics are calculated in R v3.5.2 using package vegan v2.5-3 and Stata 15 (StataCorp, Texas, USA) [Citation15].
Results
Diverticulosis
Eighty-three participants with asymptomatic diverticulosis had adequate sigmoid biopsies and they were matched with 222 controls without diverticulosis. Likewise, 37 participants with asymptomatic diverticulosis had adequate fecal samples and were matched with 104 controls without diverticulosis. Matching generated an equal distribution regarding age, sex, smoking status and recent antibiotics use, although in the fecal samples analysis the controls had a significantly lower BMI than the cases (). Also, for the analysis of mucosal biopsies there was no statistically significant difference in terms of more recent antibiotic use in cases compared with controls (p = .069; ).
Table 1. Demographic data for diverticulosis compared with non-diverticulosis.
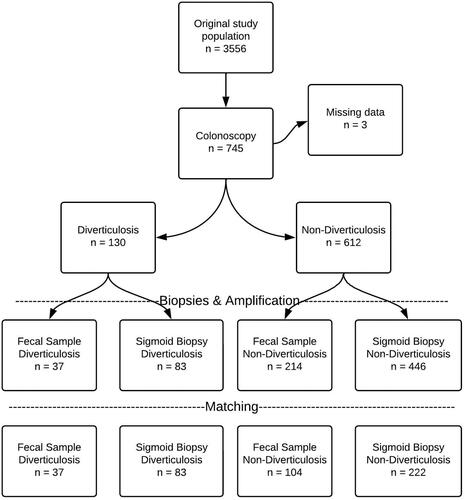
In the mucosal biopsy analysis, there was no significant difference in richness (p = .65), diversity (p = .41) or taxonomic composition (p = .39 − 0.63) ( and ). In the fecal microbiota analysis, there was no significant difference in richness (p = .28), diversity (p = .15) or taxonomic composition (p = .17 − 0.80) ( and ).
Figure 2. Richness and diversity in mucosa-associated (top two panels) and fecal microbiota (lower two panels) in individuals with diverticulosis at the time of the baseline endoscopy compared with those without diverticulosis. No significant differences were noticed in Chao 1 Richness or Shannon Diversity.
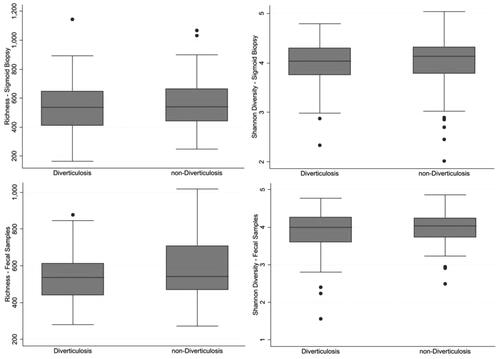
Figure 3. Taxonomic distribution in fecal and mucosa-associated microbiota in individuals with and without diverticulosis at the baseline endoscopy (top two panels) and those who went onto develop diverticulitis compared with those who did not (lower two panels) represented by Non-Metric Multidimensional Scaling (NMDS). Filled squares show cases and empty squares show controls.
Incident diverticulitis
Among the 742 individuals with information on diverticulosis status in the PopCol study, we identified 21 who later developed acute diverticulitis (21/742, 2.8%). Of the 130 individuals that had diverticulosis at the colonoscopies performed in PopCol 13 later developed acute diverticulitis (13/130, 10%). On the other hand, of the 612 participants that did not have diverticulosis at the colonoscopies eight later developed acute diverticulitis (8/612, 1.3%).
The diverticulitis cases were matched with controls that did not later develop acute diverticulitis and were matched with individuals that had the same diverticulosis status on colonoscopy. Fourteen had sigmoid biopsies that were matched with 64 controls and eight had fecal samples that were matched with 37 controls. Matching generated a similar distribution regarding age, sex, BMI, smoking status and recent antibiotic use ().
Table 2. Demographic data for participants who later developed acute diverticulitis compared with those who did not.
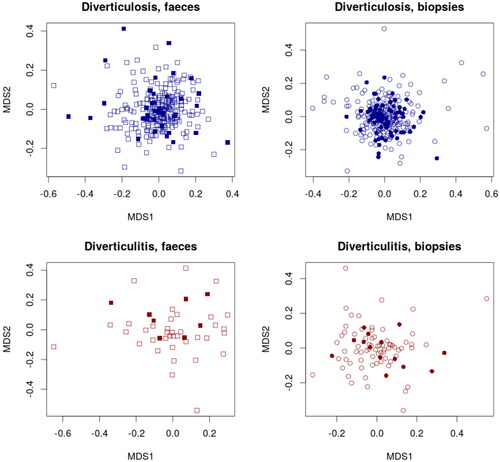
Comparing mucosal biopsies, there was no significant difference in richness (p = .22) or diversity (p = .38) (). Regarding taxonomic composition participants who later developed diverticulitis had a significantly higher abundance of genus Comamonas compared with those who did not develop diverticulitis (p = .027). Taxonomic distribution is shown in .
Figure 4. Richness and diversity in mucosa-associated (top two panels) and fecal microbiota (lower two panels) in individuals who later developed diverticulitis compared with those who did not. No significant differences were noticed in Chao 1 Richness or Shannon Diversity.
In the fecal sample analysis, there was no significant difference in richness (p = .13) or diversity (p = .069) (). No taxonomic differences were noted at the family levels (p = .07−.72) ().
Discussion
A clear role of microbiota in the pathogenesis of diverticular disease has not been established and previous studies have shown divergent results. Our population-based study on mucosa-associated and fecal microbiota did not show differences in richness, diversity or taxonomy composition between individuals with or without diverticulosis. Regarding the taxonomy composition, individuals who later developed diverticulitis had a higher abundance of the genus Comamonas compared with controls. However, this clade had a very low abundance making it difficult to assign biological significance, although bacteria in low abundance potentially can have a significant impact on colonic function and homeostasis through their metabolic activity. We did not find differences in richness or diversity between individuals who later developed diverticulitis compared with those who did not.
Few studies have compared the microbiota of individuals with diverticulosis with those without diverticulosis. Jones et al. detected a small difference in both richness, diversity and taxonomic composition where cases were more abundant for phylum Proteobacteria and genus Comamonas. Barbara et al. found that patients with diverticulosis and SUDD had a lower relative abundance of Clostridium cluster IV bacteria [Citation6]. This is in contrast with our findings that showed no significant difference in phylum Proteobacteria, genus Comamonas and Clostridium cluster IV between participants with diverticulosis compared with those without it [Citation3,Citation6]. The reason for this difference is unclear but might reflect a Type II error since the differences they found were small.
When looking at individuals who did develop acute diverticulitis Daniels et al. observed that acute diverticulitis patients have a higher diversity of fecal microbiota than controls [Citation4]. This result does not align with our outcome where there was no significant difference between the diversity in the fecal samples. Although both studies addressed acute diverticulitis, a major difference is that Daniels et al. sampled patients with ongoing acute diverticulitis whereas the current study focused on fecal samples taken before individuals developed diverticulitis, a much stronger study design. Thus, the higher diversity observed by Daniels may be a consequence of acute inflammation. Studies have found that probiotics do not lower the recurrence of acute diverticulitis which is in keeping with a lack of an association between the microbiota and future diverticulitis, corroborating the results reported here [Citation17,Citation18].
A strength of our study is the population-based design which allows for less biased case selection and this is the only population-based study on microbiota in diverticulosis and in individuals who later develop diverticulitis. In the PopCol study, the diverticular disease was rare in participants under 40 years of age at the time of investigation and the median age of the cases in the present study was about 60 years. As previously reported, any symptom selection bias was limited because we have shown that symptoms did not influence participation in the study or in undergoing colonoscopy, supporting the generalizability of the study [Citation9]. The demographic differences between individuals with and without diverticulosis reflect known risk factors for diverticulosis and DD (significantly higher BMI in the diverticulosis group) implying that the participants are representative of diverticulosis patients more widely, enhancing the generalizability of the results reported here. In other studies, participants were recruited after they had symptoms or completed a screening or surveillance colonoscopy, opening the possibility for selection bias [Citation5–8]. Furthermore, this is the first report on fecal and mucosa-associated microbiota composition in individuals before they develop diverticulitis.
One limitation of our investigation is the lack of samples from some participants and that some samples did not produce adequate microbiota data. Further, we have a relatively modest sample size, especially in the comparison of individuals who developed diverticulitis versus individuals that did not. We acknowledge that only 20% of the original population completed a colonoscopy (745/3,556). However, diverticular disease is rare in individuals under forty, and when only looking at those over forty years 29% did undergo a colonoscopy (585/2,003). Also, we did not account for proton pump inhibitor (PPI) that previously has been shown to affect the microbiota and could be a source of bias [Citation19]. However, the use of PPI was similarly low in both cases and controls, and is unlikely to have altered the findings ( and ). One limitation of all studies on this subject, including the current study, is that biopsies were not taken from inside the diverticulum which might show a different microbiota composition than random samples from outside the diverticula. However, taking a biopsy from within the diverticulum in vivo confers an unacceptable risk of complications in the form of bleeding or perforation. Future studies should investigate the use of brushes or swabs as a sampling method for the mucosal microbiota from within the diverticula.
The etiology of diverticulosis as well as acute diverticulitis is still incompletely understood, motivating further investigation. Larger population-based studies are needed to help us understand etiopathogenesis and hopefully lower the burden of this costly ailment. In this population-based cohort study the only significant difference was that those who later develop diverticulitis had more abundance of genus Comamonas. However, its biological significance is unclear suggesting overall a limited role of the gut microbiota in the etiopathogenesis of diverticular disease.
Ethical approval
The baseline study was approved by the local ethics committee at Karolinska Institutet (D.nr. 394/01). The linkage and patient file review were approved by the regional ethics board in Stockholm (D. nr. 2016/228-31/2 and 2017/603-32).
Informed consent was obtained from all participating individuals.
Disclosure statement
No potential conflict of interest was reported by the author(s)
Data availability statement
Data are available in a public, open access repository. The raw sequencing data is freely available from the European Nucleotide Archive as study PRJEB31817, samples ERS3379832-ERS3380418, available at https://www.ebi.ac.uk/ena/data/view/PRJEB31817.
Additional information
Funding
References
- Peery AF, Crockett SD, Murphy CC, et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2018. Gastroenterology. 2019;156(1):254–272.e11.
- Strate LL, Morris AM. Epidemiology, pathophysiology, and treatment of diverticulitis. Gastroenterology. 2019;156(5):1282.e1–1298.e1.
- Spiller R. Is it diverticular disease or is it irritable bowel syndrome? Dig Dis. 2012;30(1):64–69.
- Kupcinskas J, Strate LL, Bassotti G, et al. Pathogenesis of diverticulosis and diverticular disease. J Gastrointestin Liver Dis. 2019;28(Suppl 4):7–10.
- Jones RB, Fodor AA, Peery AF, et al. An aberrant microbiota is not strongly associated with incidental colonic diverticulosis. Sci Rep. 2018;8(1):4951.
- Daniels L, Budding AE, de Korte N, et al. Fecal microbiome analysis as a diagnostic test for diverticulitis. Eur J Clin Microbiol Infect Dis. 2014;33(11):1927–1936.
- Tursi A, Mastromarino P, Capobianco D, et al. Assessment of fecal microbiota and fecal metabolome in symptomatic uncomplicated diverticular disease of the Colon. J Clin Gastroenterol. 2016;50(Suppl 1): S9–S12.
- Barbara G, Scaioli E, Barbaro MR, et al. Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut. 2017;66(7):1252–1261.
- Kjellstrom L, Molinder H, Agreus L, et al. A randomly selected population sample undergoing colonoscopy: prevalence of the irritable bowel syndrome and the impact of selection factors. Eur J Gastroenterol Hepatol. 2014;26(3):268–275.
- Jarbrink-Sehgal ME, Andreasson A, Talley NJ, et al. Symptomatic diverticulosis is characterized by loose stools. Clin Gastroenterol Hepatol. 2016;14(12):1763–1770 e1.
- Hugerth LW, Andreasson A, Talley NJ, et al. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut. 2020;69(6):1076–1084.
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12.
- Edgar RC. UNOISE2: improved error-correction for illumina 16S and ITS amplicon sequencing. BioRxiv. 2016. DOI:10.1101/081257
- Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6.
- Kindt J, Gavin P, Szoecs L, et al. Vegan: community ecology package. R package version 2.5-3. 2018 [cited 2018 May 1]. Available from: https://CRAN.R-project.org/package=vegan
- Mallick H, Rahnavard A, McIver LJ, et al. Multivariable association discovery in population-scale meta-omics studies. PLOS Comput Biol. 2021;17(11):e1009442.
- Hall J, Hardiman K, Lee S, et al. The American society of colon and rectal surgeons clinical practice guidelines for the treatment of left-sided colonic diverticulitis. Dis Colon Rectum. 2020;63(6):728–747.
- Rezapour M, Ali S, Stollman N. Diverticular disease: an update on pathogenesis and management. Gut Liver. 2018;12(2):125–132.
- Bastard L, Berthelot Q, Soulillou L, et al. E. Impact of non-antibiotic drugs on the human intestinal microbiome. Expert Rev Mol Diagn. 2021;21(9):911–924.