Abstract
Little is known about the distribution of arbuscular mycorrhizal fungi (AMF, phylum Glomeromycota) in the Turkish arable soils. In this study, we investigated AM fungal phylotype composition in the roots of 13 different plant samples from one site each of the East Black Sea, Mediterranean, and Central Anatolian regions of Turkey. Fifty-seven distinguished operational taxonomic units at 97% nucleotide sequence identity were recorded among 424 partial sequences of the nuclear ribosomal large subunit (LSU) RNA genes determined. Most of the new sequences were clustered within 10 well-resolved phyloclades of the order Glomerales. About half of the newly determined sequences lacked similar sequences in the public databases. In particular, all sequences from Camellia sinensis collected in the East Black Sea region had only 83–97% sequence similarity to known AMF species. The findings suggest that novel and endemic AMF species may exist in Turkish agricultural soils. The AM fungal community composition in the East Black Sea region was relatively simple and completely differed from those in the other two regions, presumably due to the low soil pH and host specificity. The AM fungal community compositions of the Mediterranean and Central Anatolian samples were broadly similar; however, some sequences related to Rhizophagus were found only in the Mediterranean samples. This reflects the trend that more diverse AM fungal communities are established in the Mediterranean region than the Central Anatolian region.
INTRODUCTION
Arbuscular mycorrhizal fungi (AMF) are important components of natural and agricultural ecosystems, as they form symbiotic associations with the roots of most terrestrial plants. Symbiotic relationships between AMF and host plant species date from approximately 400–600 million years ago, and are thought to have a critical role in the early establishment of land plants (Remy et al. Citation1994). AMF are obligate biotrophs that belong to the phylum Glomeromycota (Schussler et al. Citation2001), and the symbioses formed between AMF and host plants are normally mutualistic (Smith and Read, Citation2008). Mycorrhizal association involves the transfer of mineral nutrients from the fungus to the host plant in exchange for carbon (C) (Smith and Read, Citation2008). In particular, it is well known that absorption of phosphorus (P) to the host plant is enhanced by AMF symbiosis (Allen Citation1991). In addition, AMF provide several other benefits to host plants, including tolerance of water deficiency, mitigation of heavy metal toxicity and protection from pathogens, as well as improvement in soil physical properties due to stabilization of soil aggregates by hyphal elongation (Allen Citation1991; Newsham et al. Citation1995; Joner et al. Citation2000; Augé Citation2001). AMF can therefore have significant positive effects on agricultural systems, and improve crop growth and productivity.
The Republic of Turkey is one of the world’s largest agricultural countries, and the fourth largest producer of vegetables according to FAOSTAT 2010. On the other hand, soil salinization, erosion and missuse, overgrazing, heavy urbanization and industrialization bring about increasing soil degradation problems in Turkey (Camci et al. Citation2007). According to UNEP (Citation1993), most areas in Turkey can be considered to suffer from desertification or have high potential for desertification. Due to climatic and topographic conditions, soil erosion is a major problem in Turkey and almost 86% of the land suffers from some degree of erosion (Ozden et al. Citation2000). Biodiversity investigations of indigenous soil microorganisms such as plant growth-promoting rhizobacteria, symbiotic bacteria and symbiotic fungi are important for the restoration of degraded soil and for sustainable agriculture.
There have, however, been only a few molecular ecological studies of AMF diversity or community composition in Turkish arable soils. By counting the AMF infection rate in the host plant roots in Turkish south Mediterranean dunes, Çakan and Karataş (Citation2006) revealed that rate of AMF association with host plants increased with progressing desertification. Karaarslan and Uyanoz (Citation2011) investigated the relationships between indigenous AMF and soil physical or chemical properties by researching AMF spore density and AMF infection rate in some native plants growing in saline soils around Lake Tuz, Turkey. They found a significant correlation between numbers of AM fungal spores and soil calcium carbonate (CaCO3) concentration, and indicated that AM fungal community structures varied to suit the stresses in saline soils. In a field experiment conducted in the Turkish Mediterranean region, Celik et al. (Citation2004) indicated that organic fertilizer or/and inoculation with AMF spores could improve soil physical and chemical properties. It was indicated that the indigenous AMF isolated from the Mediterranean region can significantly contribute to plant growth and P uptake in Turkey (Aka–Kacar et al. Citation2010; Almaca and Ortas Citation2010; Ortas Citation2010). These works have focused on AMF infection rate, spore morphology and density. On the other hand, there has been no effort to investigate AM fungal diversity through DNA analysis, as far as we know.
In the light of these facts, we aimed to elucidate the indigenous AM fungal diversity and community composition in 15 plant roots collected from three regions in Turkey, where the climatic conditions, soil characteristics and farming practices differ. Community composition was determined based on partial nucleotide sequences of the 28S large subunit ribosomal RNA genes (LSU rDNA), which were amplified from DNA extracts of fine root samples.
MATERIALS AND METHODS
Sampling sites
Sampling took place in different geographic regions of Turkey: the East Black Sea, Central Anatolia and the Mediterranean. In the East Black Sea region, samples were collected around two different locations, (i) the Atatürk Tea and Garden Research Institute located in Rize Province (41°01'N, 40°30'E, Site B1–B3), where the regional climate is semi-humid with a 15.8°C mean annual temperature, 2304.8 mm of rainfall and 77.6% humidity and (ii) a hazelnut grove located in Trabzon Province (40°54'N, 40°08'E, Site B4) where the regional climate is again semi-humid with a 15.6°C mean annual temperature, 812.0 mm of rainfall, and 71.3% humidity (TSMS Citation2009). The Central Anatolian region was represented by soil samples collected at the Kenan Evren Research Farm of Ankara University located in Haymana, Ankara Province (39°37'N, 32°31'E, Site A1–A5), where the climate is typically continental with a 13.0°C mean annual temperature, 462.2 mm of rainfall, and 60.1% humidity (TSMS Citation2009). The soil samples in the Mediterranean area were collected from various agricultural fields at the East Mediterranean Agricultural research Institute, located in Adana Province, (36°51'N, 35°20'E, Site M1–M6), where the climate is typically Mediterranean with a mean annual temperature of 19.4°C, precipitation of 811.0 mm, and 81.1% humidity (TSMS Citation2009).
According to the classification of the World Reference Base for Soil Resources (WRB), the soils at those sites were classified as Eutric Cambisol in sites B1–B4 in the East Black Sea region, Calcaric Cambisol in sites A1–A5 in Central Anatolia and Molli-Lithic Leptosol in sites M1–M6 in the Mediterranean region (Jones et al. Citation2005). Selected soil and vegetation characteristics are summarized in .
Table 1 Selected soil and vegetation characteristics
Soil and plant root sampling
Bulked samples of 500–750 g soil from the rhizosphere of three plant individuals per species were taken and passed through a 2-mm sieve. The fine roots were collected and frozen at –20°C, whereas the soil samples were stored at 4°C prior to analysis.
Soil physical and chemical properties
Soil particle size and texture were measured using the Bouyoucos hydrometer method (Bouyoucos Citation1951). Soil pH (H2O) was measured in a 1:2.5 soil:water [weight/volume (w/v)] suspension, using an electrode after shaking for 30 min. Available P (Truog-P) was measured using the vanado-molybdate method after extraction with 0.001 M sulfuric acid (H2SO4) at a ratio of 1:200 (w/v). Total C and nitrogen (N) were analyzed using an MT-700 Mark 2 CN analyzer (Yanaco, Kyoto, Japan).
Molecular analysis
The dried root samples were ground in liquid nitrogen to get a 20 mg sample for total DNA extraction, using the ISOPLANT kit (Nippon Gene, Tokyo, Japan). Part of the LSU rDNA was amplified in a 25-μL reaction, using the KOD-Plus ver. 2 polymerase chain reaction (PCR) mix (Toyobo, Osaka, Japan) with 10 μmol L–1 of each primer and 1 μL of template DNA. Nested PCR amplification rounds were carried out with the LSU rDNA-universal forward primer LR1 (van Tuinen et al. Citation1998)/the fungal LSU rDNA-specific reverse primer FLR2 (Trouvelot et al. Citation1999) for the first PCR, and the forward primer FLR3/reverse primer FLR4 (Gollotte et al. Citation2004) using 1 μL of a 1:100 dilution of the first PCR product as the template for the second PCR. A Takara PCR Thermal Cycler Dice® Gradient (Takara, Ohtsu, Japan) was used, with twice the following program: initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 96°C for 10 s, annealing at 54°C for 30 s, polymerization at 72°C for 30 s, and final elongation at 68°C for 7 min. The PCR products were cloned into the p-GEM® T-Easy Vector (Promega, Tokyo, Japan) and transformed into ECOSTM Competent Escherichia coli DH5α cells (Nippon Gene) following the manufacturer’s instructions.
For each sample, 32 recombinant bacterial colonies were randomly chosen from the DNA libraries and purified using a High Pure Plasmid Isolation Kit (Roche, Basel, Switzerland) following the manufacturer’s instructions. The nucleotide sequences were determined by dideoxy sequencing at the sequencing facilities of FASMAC Co. Ltd. (Kanagawa, Japan).
Analysis of community composition
To verify AMF identity, the sequences were compared with all known sequences in GenBank using the Basic Local Alignment Search Tool (BLAST; Altschul et al. Citation1990) on the National Center for Biotechnology Information (NCBI) website. Those sequences showing high similarity to organisms outside the phylum of Glomeromycota were excluded from subsequent analyses. The sequences were aligned in Mega 5.10 (Tamura et al. Citation2011), using ClustalW (Thompson et al. Citation1994) and adjustments were made if necessary.
To define operational taxonomic units (OTUs), a distance matrix was computed using DNADIST ver. 3.5c (J. Felsenstein, University of Washington, Seattle, WA, USA). OTUs were defined at 97% sequence identity. Potential phylotype richness (Chao1) and the associated 95% confidence interval (CI) were estimated in the Mothur software (Schloss et al. Citation2009). The Shannon diversity index was calculated for each sample with a 95% CI based in the 97% sequence identity cutoff.
An additional multiple sequence alignment was assembled in Mega 5.10 that included our new sequences and highly similar ones of formally described AMF species from GenBank. This big alignment was used to create a phylogenetic tree in Mega 5.10, using the neighbour-joining algorithm with 1000 bootstrap replicates for statistical branch support. The tree was rooted with a sequence of Paraglomus occultum. These sequence data have been submitted to the DDBJ database (accession numbers AB786346–AB786517).
To compare the AMF communities among sites, the clade type abundances and soil physical and chemical properties were submitted to a canonical correspondence analysis (CCA) using R package vegan (Oksanen et al. Citation2013).
RESULTS AND DISCUSSION
Soil chemical properties
Soil physical and chemical properties are shown in . The East Black Sea soils (B1–B4) were acidic and ranged between pH 4.47 and 5.02, while those of the Mediterranean Region (M1–M6) and Central Anatolia (A1–A5) were alkaline and ranged between pH 7.95 and 8.64.
Table 2 Soil physical and chemical properties at field sites
The pH values of the soil from Camellia sinensis varied among the fertilizer treatments; the plant compost treatment (B1), the chemical fertilizer treatment (B2), and the control (B3) had a pH of 4.65, 4.90 and 5.02, respectively. The available P, total C, and total N values of the soils in sites B1 and B2 were higher than those of the control (B3).
AMF taxa and diversity
In the present study, a total of 480 clones (32 clones per sample) were amplified, and 424 AMF clone sequences were determined (an average of 28 sequences per sample). The numbers of AMF sequences, species, OTUs (based on 97% similarity), Shannon indices, Simpson indices and estimated phylotype richness (Ace and Chao1) are shown in . A total of 169 types of AMF sequences were amplified and were ascribed to 57 OTUs.
Table 3 Absolute number of arbuscular mycorrhizal fungi (AMF) clones found at field sites and number of operational taxonomic units (OTUs), diversity indices, potential phylotype richness and coverage at a taxonomic level equating to 97% sequence similarity
Estimated phylotype richness of the samples from the Mediterranean (M1–M6) and Central Anatolian region (A1–A5) tended to be higher than those from the East Black Sea (B1–B4). The samples obtained from C. sinensis in the plant compost (B1) and the chemical fertilizer treatment (B2) were dominated by only one OTU. The coverage of OTU (OTU obtained/estimated phylotype richness) averaged 85%. Although the samples obtained from Zea mays L. (M2) in the Mediterranean had a low Shannon index, the diversity indices of the samples from this region tended to be higher than those of the other regions. Using BLAST analysis, 38 types of sequence (28 OTUs) had less than 97% sequence similarity to the known AMF sequences in the GenBank database.
No previous study has assessed the AMF assemblages in Turkish arable soils, using a molecular approach. Our 38 sequences with lower than 97% sequence similarity to the known species in the GenBank database suggest that novel and endemic AMF species may exist in Turkish arable land. These sequences were found in all samples except for C. avellana (B4) and P. armeniaca (A5) sampled in the East Black Sea and Central Anatolia, respectively. All sequences from C. sinensis in the East Black Sea had only 83–97% sequence similarity to the known AMF species.
Table 4 Clade distribution. The number of clones obtained and operational taxonomic units (OTUs) at a taxonomic level equating to 97% sequence similarity. The numbers of clones and OTUs with lower than 97% sequence similarity to known species in the GenBank database are given in parentheses
The samples of T. repens (A1) and Z. mays (A3) obtained from Central Anatolia and H. annuus (M1), Z. mays (M2), G. max (M4), M. sativa (M5) and C. sinensis (M6) from the Mediterranean involved sequences that showed high similarity (> 97%) to the AMF sequences from potato (Solanum tuberosum L.) roots in arable soils in Italy, another Mediterranean country (Cesaro et al. Citation2008).
Phylogenetic distance analysis
contains the neighbor-joining tree showing the recovered AMF phylotypes. The number of sequences and OTUs with < 97% sequence similarity, and the distribution of clades, is shown in .
Figure 1 Neighbor-joining phylogenetic tree of arbuscular mycorrhizal fungi (AMF) sequences (28S rDNA region) obtained during our study, GenBank sequences of known Glomeromycota and other GenBank AMF sequences highly similar to our sequences. Tree is rooted to Paraglomus occultum (FJ461883). Bootstrap values of 70% (1000 replicates) and higher are indicated. Rhi: Rhizophagus, Fun: Funneliformis, Cla: Claroideoglomus, Aca: Acaulospora, Unc: Uncultured glomeromycota.
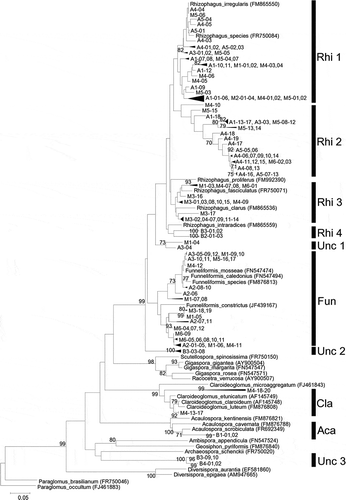
Ten distinct clades are visible in the phylogenetic tree (). Clades Rhi4, Unc2, Aca and Unc3 comprised sequences from the East Black Sea region only. Clade Rhi3 and Cla comprised sequences from the Mediterranean only. Clades Rhi1, Rhi2, Unc1, and Fun comprised sequences from the Mediterranean and Central Anatolia.
The sequences obtained from C. sinensis in the plant compost (B1) and chemical fertilizer treatments (B2) formed different clades from each other (). The sequences found in the plant compost treatment were related to Acaurospora (clade Aca) and those in the chemical fertilizer treatment were related to Rhizophagus (clade Rhi4). Clade Rhi4 comprised AMF obtained from C. sinensis only (B2 and B3), which were related to Rhizophagus; however, all sequences in clade Rhi4 had low sequence homologies to known species (< 93% similarity). Clade Aca was only from C. sinensis in the plant compost treatment (B1) and was related to Acaulospora. These clade Aca sequences also had low sequence homologies to known species (87% similarity). Clade Unc3 was formed from Corylus avellana (B4) and C. sinensis in the control treatment (B3), and their sequences were related to the AMF species recorded as uncultured glomeromycetes (Verbruggen et al. Citation2010).
As shows, there was a significant difference in soil chemical properties (i.e. available P, total C and N) between the control site (B3) and the two fertilized sites (B1 and B2). The result means that fertilization can drastically decrease AM fungal diversities associated with C. sinensis. This is in accordance with the opinions of Singh et al. (Citation2008), who reported that various cultural practices negatively affect AMF diversity at the genus level, in tea plantations in the colder regions. They also showed that the Shannon-Weaver diversity index at the species level was higher in the natural ecosystem than in the cultivated site.
Clades Rhi2 and Fun had various sequences with < 97% sequence similarity to known species in the GenBank database (). Clade Rhi2 comprised uncultured Glomeromycota which had been previously identified by sequence analysis (Cesaro et al. Citation2008), but no spore morphology data have been available for these species.
The clade distribution among the Mediterranean and Central Anatolian sequences was broadly similar. However, as clade Rhi3 consisted of only AMF from the Mediterranean, this region contained more various AMF species related to Rhizophagus than did Central Anatolia. Considering that no significant differences were found in the soil chemical properties between these regions, climate and/or parent soil materials may affect AM fungal diversities.
In this study, the samples collected in the East Black Sea region formed completely different AM fungal communities from those in the other two regions. As shown in , there was no common clade between the samples from the East Black Sea region and those in the others. Therefore, the samples from the Black Sea region were excluded from CCA. CCA was performed to investigate differences among samples or regions based on the clade type abundances and soil physical and chemical properties among the samples from the Mediterranean and Central Anatolia. As a result, AM fungal communities from M. pumila (A4) and P. armeniaca (A5) whose soils were characterized by a high amount of total C were plotted at points near each other (). It was implied that total C affected clade Rhi2 which was dominant in samples from M. pumila (A4) and P. armeniaca (A5). It was indicated that clade Rhi3 and Cla have no relation between available P, total C or N (). It is suggested that clade Rhi3, which was comprised only from the samples collected in the Mediterranean region, shows the regional difference. In addition, clade Cla, which was obtained from G. max (M4), may reflect host specificity of AM fungi. The effect of available P on difference of AM fungal communities among the samples was small compared with total C and N.
Figure 2 Canonical correspondence analysis (CCA) of arbuscular mycorrhizal fungal communities (triangles = the Mediterranean region [M1–M6]; diamonds = the Central Anatolia region [A1–A5]). Abbreviations of arbuscular mycorrhizal fungal clades and soil physical/chemical properties correspond to and , respectively.
![Figure 2 Canonical correspondence analysis (CCA) of arbuscular mycorrhizal fungal communities (triangles = the Mediterranean region [M1–M6]; diamonds = the Central Anatolia region [A1–A5]). Abbreviations of arbuscular mycorrhizal fungal clades and soil physical/chemical properties correspond to Fig. 1 and Table 2, respectively.](/cms/asset/a2b397a2-8505-4307-a4d3-c173d1733936/tssp_a_890916_f0002_b.gif)
Uniqueness of AMF in the East Black Sea region
Why might quite unique AM fungal communities exist in the East Black Sea region as compared to the other two regions? The soil samples collected from the East Black Sea were characterized by low pH (4.62–5.02; ). While host plant composition is known to be a main driving force for structuring AM fungal communities (Johnson et al. Citation1992), alteration in AM fungal communities due to low soil pH in acid sulfate soil has also been reported (An et al. Citation2008). The difference in the AM fungal community composition found in the three Turkish soils is speculated to be due to the acidic soil pH in the East Black Sea region.
In acidic soil, low availability of P, owing to low native P content and high P fixation capacity in acidic soil is known to be one of the main limiting factors for the productivity of tea leaves (Lin et al. Citation1992). In addition, plants growing on acidic soil are often exposed to relatively severe mineral stress, including toxicity (e.g. aluminium, manganese) and deficiency (e.g. P, potassium, magnesium), as described by Marschner (Citation1991). Excess aluminium, in particular, impairs root elongation, resulting in plant growth depression in acidic soils (Foy Citation1992). The AMF-root symbiosis may help alleviate some of the problems that plants encounter when grown in acidic soils (Marschner Citation1991; Sieverding Citation1991), including Al and Mn toxicity (Maddox and Soileau Citation1991; Sieverding Citation1991). The unique AM fungal species we found in the East Black Sea region in this study are probably adapted to the low pH environment and may contribute to plant growth in this area.
CONCLUSION
We recorded the phylogenetic composition of the AMF communities in the roots of 13 different plant species collected at three sites in three regions of Turkey. This research showed the first DNA sequence data of AMF from Turkey, half of which appear to have no close similarity (> 97%) to the sequence accessions in the public databases. The AMF communities of the soils from the East Black Sea region with a distinctively lower pH than the other soils separated from all the other AMF assemblages. All sequences from C. sinensis in the East Black Sea region had only 83–97% sequence similarity to known AMF species. Although there was no significant difference of the AM fungal community compositions between the Mediterranean and Central Anatolian samples, some sequences related to Rhizophagus (clade Rhi3) indicated regional differences.
REFERENCES
- Aka–Kacar Y, Akpinar C, Agar A, Yalcin–Mendi Y, Serce S, Ortas I 2010: The effect of mycorrhiza in nutrient uptake and biomass of cherry rootstocks during acclimatization. Rom. Biotechnol. Lett., 15(3), 246–252.
- Allen MF 1991: The Ecology of Mycorrhizae. Cambridge University Press, New York, NY. pp.184.
- Almaca A, Ortas I 2010: Growth response of maize plants (Zea mays L.) to wheat and lentil pre–cropping and to indigenous mycorrhizae in field soil. Span. J. Agric. Res., 8, 131–136. doi:10.5424/sjar/201008S1-1232
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990: Basic local alignment search tool. J. Mol. Biol., 215, 403–410.
- An GH, Miyakawa S, Kawahara A, Osaki M, Ezawa T 2008: Community structure of arbuscular mycorrhizal fungi associated with pioneer grass species Miscanthus sinensis in acid sulfate soils: Habitat segregation along pH gradients. Soil Sci. Plant Nutr., 54, 517–528. doi:10.1111/j.1747-0765.2008.00267.x
- Augé RM 2001: Water relations, drought and vesicular–arbuscular mycorrhizal symbiosis. Mycorrhiza, 11, 3–42. doi:10.1007/s005720100097
- Bouyoucos GJ 1951: A recalibration of the hydrometer method for making mechanical analysis of soils. Agron. J., 43, 434–439. doi:10.2134/agronj1951.00021962004300090005x
- Çakan H, Karataş Ç. 2006: Interactions between mycorrhizal colonization and plant life forms along the successional gradient of coastal sand dunes in the eastern Mediterranean, Turkey. Ecol. Res., 21, 301–310. doi:10.1007/s11284-005-0134-x
- Camci SC, Karaca A, Haktanır K, Yildiz H 2007: Global attention to Turkey due to desertification. Environ. Monit. Assess., 128, 489–493. doi:10.1007/s10661-006-9342-2
- Celik I, Ortas I, Kilic S 2004: Effects of compost, mycorrhiza, manure and fertilizer on some physical properties of a Chromoxerert soil. Soil Till. Res., 78, 59–67. doi:10.1016/j.still.2004.02.012
- Cesaro P, van Tuinen D, Copetta A, Chatagnier O, Berta G, Gianinazzi S, Lingua G 2008: Preferential colonization of Solanum tuberosum L. roots by the fungus Glomus intraradices in arable soil of a potato farming area. Appl. Environ. Microbiol., 74(18), 5776–5783. doi:10.1128/AEM.00719-08
- Foy CD 1992: Soil chemical factors limiting plant root growth. Adv. Soil Sci., 19, 87–149.
- Gollotte A, van Tuinen D, Atkinson D 2004: Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza, 14(2), 111–117. doi:10.1007/s00572-003-0244-7
- Johnson NC, Tilman D, Wedin D 1992: Plant and soil controls on mycorrhizal fungal communities. Ecology, 73, 2034–2042. doi:10.2307/1941453
- Joner EJ, Briones R, Leyval C 2000: Metal–binding capacity of arbuscular mycorrhizal mycelium. Plant Soil, 226, 227–234. doi:10.1023/A:1026565701391
- Jones A, Montanarella L, Jones R 2005: Soil Atlas of Europe. European Soil Bureau Network. European Commision, p.128.
- Karaarslan E, Uyanoz R 2011: Occurrence of arbuscular mycorrhizal fungi in some native plants grown on saline soils around the lake Tuz in Turkey and its relations with some physical and chemical properties of soil. Sci. Res. Essays, 20, 4238–4245.
- Lin Z, Wu X, Wang X, Yu Y 1992: Studies on phosphorus nutrition in red soil of tea field. In: Tea Science research Proceedings. Tea Research Institute of Chinese Academy of Agricultural Science. Shanghai Scientific and Technological Publisher, Shanghai.
- Maddox JJ, Soileau JM 1991: Effect of phosphate fertilization, lime amendments and inoculation with VA–mycorrhizal fungi on soybeans in an acid soil. Plant Soil, 134, 83–93.
- Marschner H 1991: Mechanisms of adaptation of plants to acid soils. Plant Soil, 134, 1–20.
- Newsham KK, Fitter AH, Watkinson AR 1995: Arbuscular mycorrhiza protect an annual grass from root pathogenic fungi in the field. J. Ecol., 83, 991–1000. doi:10.2307/2261180
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH and Wagner H 2013: vegan: Community Ecology Package. R package version 2.0-7. URL http://CRAN.R-project.org/package=vegan
- Ortas I 2010: Effect of mycorrhiza application on plant growth and nutrient uptake in cucumber production under field conditions. Span. J. Agric. Res., 8, 116–122. doi:10.5424/sjar/201008S1-1230
- Ozden DM, Dursun H, Sevinc AN 2000: The land resources of Turkey and ativities of general directorate of rural services. Proceedings of international symposium on desertification, 13–17 June 2000, Konya, Turkey, p.1–13.
- Remy W, Taylor TN, Hass H, Kerp H 1994: Four hundred–million–year–old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci., 91, 11841–11843. doi:10.1073/pnas.91.25.11841
- Schloss PD, Westcott SL, Ryabin T, Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. 2009: Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol., 75(23), 7537–7541. doi:10.1128/AEM.01541-09
- Schussler A, Schwarzott D, Walker D 2001: A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycol. Res., 105, 1413–1421.
- Sieverding E 1991: Vesicular–Arbuscular Mycorrhiza Management in Tropical Agrosystems, Deutsche Gesellschaft Technische Zusammenarbeit (GTZ), Eschborn.
- Singh S, Pandey A, Chaurasia B, Palni LMS 2008: Diversity of arbuscular mycorrhizal fungi associated with the rhizosphere of tea growing in ‘natural’ and ‘cultivated’ ecosites. Biol. Fert. Soils, 44(3), 491–500. doi:10.1007/s00374-007-0231-9
- Smith SE, Read DJ 2008: Mycorrhizal Symbiosis 3rd ed. Academic Press, New York, NY.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S 2011: MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., 28, 2731–2739. doi:10.1093/molbev/msr121
- Thompson JD, Higgins DG, Gibson TJ 1994: CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. doi:10.1093/nar/22.22.4673
- Trouvelot S, van Tuinen D, Hijri M, Gianinazzi-Pearson V 1999: Visualization of ribosomal DNA loci in spore interphasic nuclei of glomalean fungi by fluorescence in situ hybridization. Mycorrhiza, 8, 203–206. doi:10.1007/s005720050235
- TSMS, Turkish State Meteorological Service 2009: http://www.meteor.gov.tr.
- UNEP 1993: World atlas of desertification: Edward Arnold.
- van Tuinen D, Jacquot E, Zhao B, Gollotte A, Gianinazzi-Pearson V 1998: Characterization of root colonization profiles by a microcosm community of arbuscular mycorrhizal fungi using 25S rDNA–targeted nested PCR. Mol. Ecol., 7(7), 879–887. doi:10.1046/j.1365-294x.1998.00410.x
- Verbruggen E, Röling WFM, Gamper HA, Kowalchuk GA, Verhoef HA, van der Heijden MGA 2010: Positive effects of organic farming on below-ground mutualists: Large-scale comparison of mycorrhizal fungal communities in agricultural soils. New Phytol., 186(4), 968–979. doi:10.1111/j.1469-8137.2010.03230.x
