Real-time PCR quantification showed that the numbers of methanogenic archaeal 16S rRNA genes and mcrA genes, encoding the methyl-coenzyme M reductase α subunit of methanogenic archaea, ranged from 4.0 × 106 to 2.7 × 108 and from 4.7 × 105 to 3.2 × 107 g−1 dry soil, respectively.
In the original version of this paper, published in Soil Science and Plant Nutrition, http://dx.doi.org/10.1111/j.1747-0765.2010.00521.x, the abstract on page 831 was incorrect. It should read:
Real-time PCR quantification showed that the numbers of methanogenic archaeal 16S rRNA genes and mcrA genes, encoding the methyl-coenzyme M reductase α subunit of methanogenic archaea, ranged from 4.0 × 106 to 2.7 × 108 and from 4.5 × 106 to 3.1 × 108 g−1 dry soil, respectively.
The numbers of targeted 16S rRNA genes and mcrA genes in the 15 paddy field soils in NE China estimated by the real-time PCR ranged from 4.0 × 106 g−1 dry soil to (BL-III) to 2.7 × 108 g−1 dry soil (BL-I) and from 4.7 × 105 g−1 dry soil to (BL-III) to 3.2 × 107 g−1 dry soil (BL-I), respectively (). The numbers of methanogenic archaea in Japanese paddy field soils of chemical plots in Anjo and Chikugo in 2003 and 2005 were 5.9 × 107– 4.5 × 108 g−1 dry soil for 16S rRNA genes and 6.1 × 106– 1.2 × 108 g−1 dry soil for mcrA genes.”
In the original version of this paper, published in Soil Science and Plant Nutrition, http://dx.doi.org/10.1111/j.1747-0765.2010.00521.x, the Quantification of methanogenic archaeal numbers on page 834 was incorrect. It should read:
The numbers of targeted 16S rRNA genes and mcrA genes in the 15 paddy field soils in NE China estimated by the real-time PCR ranged from 4.0 × 106 g−1 dry soil to (BL-III) to 2.7 × 108 g−1 dry soil (BL-I) and from 4.5 × 106 g−1 dry soil to (BL-III) to 3.1 × 108 g−1 dry soil (BL-I), respectively (). The numbers of methanogenic archaea in Japanese paddy field soils of chemical plots in Anjo and Chikugo in 2003 and 2005 were 5.9 × 107– 4.5 × 108 g−1 dry soil for 16S rRNA genes and 5.7 × 107– 1.2 × 109 g−1 dry soil for mcrA genes.
The numbers of methanogenic archaea in 15 paddy field soils in NE China were 4.0 × 106–2.7 × 108 g−1 dry soil for 16S rRNA genes and 4.7 × 105 to 3.2 × 107 g−1 dry soil for mcrA genes.
In the original version of this paper, published in Soil Science and Plant Nutrition, http://dx.doi.org/10.1111/j.1747-0765.2010.00521.x, the conclusion on page 837 was incorrect. It should read:
The numbers of methanogenic archaea in 15 paddy field soils in NE China were 4.0 × 106–2.7 × 108 g−1 dry soil for 16S rRNA genes and 4.5 × 106 to 3.1 × 108 g−1 dry soil for mcrA genes.
Corrections have been made to and according to revised information:
Table 1 Locations, soil properties and numbers of methanogenic archaea in the 15 paddy fields used in the present study
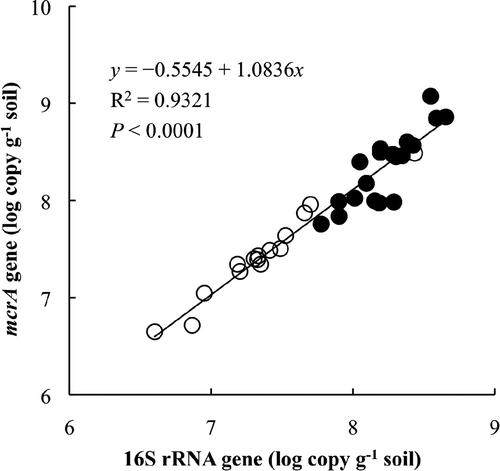
Figure 3 Relationship of quantifying methanogenic archaea by real-time polymerase chain reaction between targeting 16S rRNA genes and mcrA genes. The white circles indicate paddy field soil samples in north-east China and the black circles indicate soil samples from Japanese paddy fields of Anjo and Chikugo.
These corrections do not influence the main conclusions of this study, but invalidate a part of the discussion about the ratio of methanogenic archaeal 16S rRNA and mcrA genes at the 4th paragraph in p. 836 because the number of mcrA genes was equivalent to the number of 16S rRNA genes.
The results of the real-time PCR analysis showed that the number of methanogenic archaeal 16S rRNA genes is greater than the number of mcrA genes (). Given that 16S rRNA genes can be present in multiple copies in the bacterial chromosome (Masco et al. 2007) and that some sequences of 16S rRNA genes obtained in the present study were not derived from methanogenic archaea, the interpretation of quantitative methanogenic archaea obtained by real-time PCR might be compromised. Therefore, it is necessary to analyze the relationship between the numbers of 16S rRNA genes and mcrA genes. All data observed in NE China and in Japanese paddy field soils in Anjo and Chikugo were used for the correlation analysis, and the result clearly showed a significant linear positive relationship between the two genes, irrespective of the sampling location (). This finding suggests that both genes are suitable for quantification of methanogenic archaea, although quantification of 16S rRNA genes outnumbered that of mcrA genes.
This fact suggests that the multiple copies of 16S rRNA genes in chromosome and amplification of some non-methanogenic archaeal 16S rRNA genes did not significantly affect the estimation of methanogenic archaeal population in the paddy field soils.
