ABSTRACT
Iron (Fe) deficiency is a serious agricultural problem, especially in calcareous soils, which are distributed worldwide. Poplar trees are an important biomass plant, and overcoming Fe deficiency in poplars will increase biomass productivity worldwide. The poplar Fe-deficiency response and the genes involved in poplar Fe homeostasis remain largely unknown. To identify these genes and processes, we cultivated poplar plants under Fe-deficient conditions, both in calcareous soil and hydroponically, and analyzed their growth rates, leaf Soil and Plant Analyzer Development (SPAD) values, and metal concentrations. The data clearly showed that poplars have notable growth defects in both calcareous soil and a Fe-deficient hydroponic culture. They exhibited serious chlorosis of young leaves after 3 weeks of Fe-deficient hydroponic culture. The Fe concentrations in old leaves with high SPAD values were markedly lower in Fe-deficient poplars, suggesting that poplars may have good translocation capability from old to new leaves. The Zn concentration in new leaves increased in Fe-deficient poplars. The pH of the hydroponic solution decreased in the Fe-deficient culture compared to the Fe-sufficient culture. This finding shows that poplars may be able to adjust the pH of a culture solution to better take up Fe. We also analyzed the expression of Fe homeostasis-related genes in the roots and leaves of Fe-sufficient and Fe-deficient poplars. Our results demonstrate that PtIRT1, PtNAS2, PtFRO2, PtFRO5, and PtFIT were induced in Fe-deficient roots. PtYSL2 and PtNAS4 were induced in Fe-deficient leaves. PtYSL3 was induced in both Fe-deficient leaves and roots. These genes may be involved in the Fe uptake and/or translocation mechanisms in poplars under Fe-deficient conditions. Our results will increase a better understanding of the Fe-deficiency response of poplars and hence improve the breeding of Fe-deficiency-tolerant poplars for improved biomass production, the greening of high pH soils, and combatting global warming.
1. Introduction
1.1. Potential of the poplar as a biomass plant
Poplars (Populus spp.) are perennial trees planted for landscape and agricultural purposes worldwide. Poplar wood is used to build homes, make tools and medicine, and protect riverbanks, and they are planted for windbreaks and shelterbelts. Poplars are known as ‘the trees of the people’ and are considered one of the most important families of woody plants in terms of human use (Gordon Citation2001). Use of poplars is currently expanding due to their environmental benefits, such as phytoremediation, soil carbon sequestration, sediment run-off reduction, soil quality improvement, and wildlife habitat (Stanton et al. Citation2002).
Additionally, poplars can also be widely used for bioenergy. Poplar trees have a considerable potential for use in biofuel production (Townsend et al. Citation2014). Poplars can produce greater amounts of energy and are estimated to replace more gasoline and diesel than corn, soybeans, reed canary grass, and switchgrass (Adler et al. Citation2007). Poplar trees can produce approximately 11 barrels of oil per acre per year (Isebrands et al. Citation1979). Because of their fast growth, ability to produce a large quantity of biomass in a short period of time, and high cellulose and low lignin contents, poplars are more desirable for biofuels than many other woody crops. Moreover, poplars can be harvested throughout the year and thus provide a more continuous supply of feedstock to a bio-refinery compared with other bioenergy crops. Therefore, it is estimated that poplars have the potential to be an important component of the solution to economic and environmental problems in the twenty-first century as both human populations and greenhouse gas concentrations rise. Recently, many researchers have focused on the improvement of poplars for bioenergy, carbon sequestration, phytoremediation, and watershed protection (Gordon Citation2001).
1.2. Iron deficiency and iron uptake system in higher plants
Iron (Fe) deficiency is a widespread agricultural problem that is common in calcareous soils, which cover more than 30% of the earth’s surface. Although Fe is abundant in soil, it is found primarily in its oxidized, low-solubility form Fe (III), which is not readily available to plants, especially at the high pH of calcareous soil, under aerobic conditions (Marschner Citation1995). Under Fe-deficient conditions, plants exhibit symptoms of chlorosis in leaves and growth defects, leading to reduced crop yield and quality. Thus, Fe is a key determinant of biomass production and plant product quality (Briat et al. Citation2015). Cultivation of high-biomass crops tolerant of low Fe availability in calcareous soil could reduce the carbon dioxide concentration in the atmosphere and, consequently, ameliorate global warming.
Higher plants use two major Fe uptake strategies, known as Strategy I and Strategy II (Römheld and Marschner Citation1986). Under conditions of low Fe availability, non-graminaceous plants use Strategy I and graminaceous plants use Strategy II. Strategy I involves induction of three major processes. First, the H+-ATPase (AHA2 in Arabidopsis) acidifies the rhizosphere of Fe-deficient plants, facilitating Fe solubilization (Santi and Schmidt Citation2009). ABCG37 (AtABCG37/AtPDR9 in Arabidopsis) participates in secretion of coumarin-phenolic-type compounds from root under Fe-deficiency condition in order to chelate and mobilize Fe3+ (Fourcroy et al. Citation2014; Sisó-Terraza et al. Citation2016). The oxoglutarate-dependent oxidase feruloyl-CoA 6′-hydroxylase1 (F6′H1) is known to be involved in coumarin biosynthesis (Schmid et al. Citation2014). Recently, Rajniak et al. (Citation2018) identified two enzymes (S8H and CYP82C4) that complete the biosynthetic pathway of the coumarins; fraxetin and sideretin. Second, Fe (III)-chelate reductase (FRO) is induced in roots and reduces Fe (III)-chelate to Fe2+ (Robinson et al. Citation1999), which is assumed to be the rate-limiting step for acquisition of Fe from soil (Connolly et al. Citation2003). Third, the Fe2+ is absorbed via the Fe-regulated transporter (IRT) (Eide et al. Citation1996), which is the major Fe2+ transporter in plant roots.
FER was identified through cloning as a transcription factor related to Fe homeostasis in tomatoes (LeFER: Ling et al. Citation2002). Then, FER-like Fe-deficiency-induced transcription factor (FIT), a functional orthologue of FER, was cloned in Arabidopsis. FIT is expressed only in the roots, where it upregulates Fe uptake-related genes, such as FRO2 and IRT1 (Colangelo and Guerinot Citation2004; Jakoby et al. Citation2004; Yuan et al. Citation2005). AtFIT, also named AtbHLH29, belongs to the III(a–c) bHLH subfamily among Basic–Helix–Loop–Helix proteins in plants (Pires and Dolan Citation2010). Yin et al. (Citation2014) reported MxFIT as FIT gene of apple genus (Malus xiaojinensis). Nicotianamine (NA), a chelator of metal cations, such as Fe2+ and Zn2+, is biosynthesized from S-adenosyl methionine by NA synthase (NAS; Higuchi et al. Citation1994). All higher plants synthesize and utilize NA for the internal transport of Fe and other metals (Hell and Stephan Citation2003; Takahashi et al. Citation2003). Indeed, the NA-defective tomato mutant chloronerva (Rudolph et al. Citation1985) exhibits a phenotype indicative of Fe deficiency (Pich and Scholz Citation1996; Stephan et al. Citation1996).
In Strategy II, graminaceous plants produce mugineic acids (MAs) and secrete from the roots, which chelate Fe (III) in the soil and absorb Fe (III)-MAs complexes into the root through Fe (III)-MAs transporters called yellow stripe 1 in maize (Curie et al. Citation2001). Koike et al. (Citation2004) identified 18 putative Fe (III)-MAs transporter genes in rice genome and showed that OsYSL2 transported ferrous Fe(II)-NA and manganese-NA. OsYSL2 is preferentially expressed in leaf phloem cells and vascular bundles of flowers and developing seeds, suggesting a role in internal Fe transport. In contrast, OsYSL15 functions in the rice root cells to uptake Fe (III)-MAs from the soil (Inoue et al. Citation2009).
YSLs are also found in non-graminaceous plants and YSLs in non-graminaceous plants have been hypothesized to transport metals complexed by NA. AtYSL1 is essential for Fe (II)-NA seed loading (Jean et al. Citation2005). AtYSL2 was reported to be an Fe (II)-NA and copper-NA transporter (DiDonato et al. Citation2004).
1.3. Fe deficiency and the Fe uptake system in poplars
There have been some reports of the mechanisms of metal transport in poplars. Fe-deficiency problems have been observed in European cultivated poplars (Keller and Koch Citation1964). The genome sequence of Populus trichocarpa was deciphered by Tuskan et al. (Citation2006). The gene expression levels of metal transporters in the zinc-regulated transporter, iron-regulated transporter-like protein, natural resistance-associated macrophage protein, and heavy metal ATPase families have been reported in poplars grown under normal cultivation conditions (Migeon et al. Citation2010). Poplar YSL transporters were characterized by Cao et al. (Citation2011). PtIRT1 and PtIRT3 were reported in poplars by Huang and Dai (Citation2015), who used a yeast complementation assay to confirm that PtIRT1 and PtIRT3 are ferric transporters and showed that PtIRT1 was induced in Fe-deficient roots. However, a detailed analysis of Fe-deficiency response genes, such as YSLs, NASs, FROs, and FITs, has not yet been performed in poplars under Fe-deficient conditions. Moreover, there are still no reports of the relationships among growth defects due to Fe deficiency, metal concentration, and gene expression in poplar roots and shoots.
Desert greening has emerged as a promising approach to fighting global warming in this century. Poplars are suitable biomass plants (Stanton et al. Citation2002), with good cold resistance (Sakai and Larcher Citation1987). Moreover, poplars are very suitable for desert greening in northwestern China and Mongolia. For example, Seiei Toyama used poplars for greening the deserts in Mongolia (Ramon Magsaysay Award for Peace and International Understanding, Citation2003). Mongolia has high-pH soils (6.5–9.5) (Kobayashi et al. Citation2012). Calcareous soils or high-pH soils are widespread in desert areas (FAO-GIS Citation1998; IGBP-DIS Citation1998). Thus, breeding poplars with tolerance to Fe deficiency are highly desirable for enhancing desert greening efforts.
1.4. Purpose of this study
The aims of this study were to comprehensively analyze growth defects, symptoms, and Fe distribution in the leaves of poplars and to identify changes in gene expression in response to Fe deficiency for future breeding of poplars with tolerance to Fe deficiency. This study involved the physiological and expression analysis of Fe homeostasis-related genes using hydroponically grown poplar seedlings exposed to Fe-deficient and Fe-sufficient conditions. Poplars were also cultured on normal soil and calcareous soil, and growth under each treatment condition was analyzed. We carried out a growth experiment and metal concentration analysis and determined various gene expression patterns in the roots and leaves of Fe-deficient and Fe-sufficient poplars.
2. Materials and methods
2.1. Plant cultivation conditions in soil
The poplar plants (hybrid aspen: Populus tremula × Populus tremuloides, T89) used in this research were cultivated and propagated under aseptic conditions on 1/2 Murashige and Skoog medium without sucrose. Poplar seedlings (10 cm in height) were grown on 750 cm3 of peat moss-based soil (Jiffy-Mix; Sakata Seed Co. Ltd., Tokyo, Japan) mixed with 250 cm3 of vermiculite (vs-kakou Co. Ltd., Tokyo, Japan) under normal or control conditions or on 750 cm3 of calcareous soil obtained from Takaoka City, Toyama, Japan (Nihonkai Kougyou, Japan; pH 8.9, soluble CaO: 39.6%, Fe2O3: 1.7%) mixed with 250 cm3 of vermiculite under Fe-deficient conditions. HYPONeX (NPK: 6–10-5, Mn 13 ppm, Fe 490 ppm, Cu 3.86 ppm, and Zn 6.11 ppm; HYPONeX JAPAN Co. Ltd., Osaka, Japan) solution was used as a fertilizer. HYPONeX fertilizer was diluted 2000 times with ion-exchanged water, and 250 ml was applied every week. Plant heights were measured throughout cultivation. The estimated sizes of the fourth, fifth, and sixth newest leaves at 18 days after transplanting to normal or calcareous soil were calculated by the following formula:
2.2. Plant cultivation conditions in hydroponic culture
Poplar seedlings (P. tremula × P. tremuloides, T89), 10 cm in height, were grown in hydroponic culture solution (modified Hoagland solution) containing 5.0 mM KNO3, 5.0 mM Ca(NO3)2•4H2O, 91.4 μM Fe-EDTA, 2.0 mM MgSO4•7H2O, 1.0 mM NH4NO3, 1.0 mM KH2PO4, 46.3 μM H3BO3, 9.17 μM MnSO4•H2O, 0.765 μM ZnSO4•7H2O, 0.20 μM CuSO4•5H2O, and 0.50 μM Na2MoO4•2H2O at pH 5.6. Aeration was provided by a commercial air pump in a growth chamber maintained at 20°C under 300 µmol m−2 s−1 fluorescence light during 16 h (day) and at 18°C with no light during 8 h (night), with constant 80% humidity. The pH of the solution was adjusted to 5.6 every 2 or 3 days, and the culture solution was renewed every 7 days throughout cultivation. After 17 days, poplar plants grown in the hydroponic culture were moved from the growth chamber to a greenhouse under a schedule of 30°C for 14 h (day) with supplemental fluorescent lighting and 25°C for 10 h (night) under natural lighting conditions.
In terms of the Fe-deficient treatment, after an additional 10 days, roots were thoroughly washed with MilliQ water and 20 mM EDTA-Na, and plants were then transplanted to new hydroponic culture solutions with or without Fe for continued growth in the greenhouse. Then, leaves of 54-day Fe-deficient or Fe-sufficient-treated plants were harvested for metal concentration analysis (first hydroponic cultivation). Sixty-one-day Fe-deficiency-treated plants were transferred new hydroponic culture solution with Fe and phenotypes were observed (first hydroponic cultivation). Next, poplar seedlings were cultivated in the hydroponic culture conditions described above as second hydroponic cultivation. Four biological replicates (n = 4) were used for each Fe condition. Plant heights and main trunk diameters (at 10 cm in height from the cover of the culture box) were measured throughout cultivation. Fe-deficiency-related chlorosis of leaves was also examined through the Soil and Plant Analyzer Development (SPAD) value obtained with a SPAD-502 Plus chlorophyll meter (KONICA MINOLTA, Tokyo, Japan). After 10 days of exposure to Fe-deficient conditions, roots and the fifth newest leaves were sampled for RNA extraction and gene expression analysis by real-time reverse transcription polymerase chain reaction (real-time RT-PCR) (Fig. S3). After 37 days, leaves were harvested and scanned to image and then leaf area was measured by AR_CAD software (SHF Co. Ltd. Japan).
2.3. Metal concentration analysis
New leaves (with SPAD value around 5), middle leaves (with SPAD value around 20), and old leaves (the leaves that exist since before Fe-deficiency treatment; with SPAD value around 40) from 54-day Fe-deficient hydroponically grown plants and new or old leaves that were in the corresponding position from Fe-sufficient hydroponically grown plants were collected for measurement of metal concentrations. Dried samples were digested with 2 ml HNO3 (Wako, Japan) and 2 ml H2O2 (Wako, Japan) using the MARS Xpress digester (CEM Japan) at 220°C for 20 min and filtered as described by Masuda et al. (Citation2009). We measured the concentrations of Fe, zinc (Zn), manganese (Mn), copper (Cu), and calcium (Ca) in the digested samples using an inductively coupled plasma atomic emission spectrometer (ICPS-8100; Shimadzu, Kyoto, Japan) according to the method of Masuda et al. (Citation2009) using three biological replicates (n = 3).
2.4. Gene expression analysis through real-time RT-PCR
Quantitative real-time RT-PCR analysis was performed to determine the expression patterns of Fe homeostasis-related genes. RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN, Tokyo, Japan) using a modified method for poplar plants (Brunner et al. Citation2004). First-strand cDNA was synthesized using the ReverTra Ace kit with genomic DNA remover (Toyobo, Osaka, Japan). Gene-specific primers were designed as follows: transcript sequences of PtYSLs were obtained from Cao et al. (Citation2011) and the NCBI database. To obtain transcript sequences of PtNASs, PtFROs, and PtFIT and to design primers for real-time RT-PCR, the amino acid sequences of Arabidopsis NAS (AtNAS1, accession no. At5g04950), Arabidopsis FRO (AtFRO2, accession no. At1g01580), and Arabidopsis FIT (AtFIT, accession no. At2g28160) were used to query the Basic Local Alignment Search Tool (BLAST) against the whole-genome sequence of P. trichocarpa using the Populus Genome Integrative Explorer (http://popgenie.org). The amino acid sequence of Arabidopsis PDR9 (At3g53480), F6′H1 (At3g13610), S8H (At3g12900), CYP82C4 (At4g31940), and AHA2 (At4g30190) was also used to find poplar homologue by the same process described above. PtTIF5α, encoding poplar translation initiation factor eIF5, was used as an internal control gene (Huang and Dai Citation2015). For PtIRT1, PtIRT3, and PtTIF5α, primer sequences reported by Huang and Dai (Citation2015) were used. The accession numbers of the genes used are listed in Supplemental Table 1.
Quantitative real-time RT-PCR was performed using the StepOnePlus™ Real-Time PCR System (Life Technology, Tokyo, Japan) with SYBR Premix Ex Taq II reagent (Takara, Shiga, Japan). Transcript levels were normalized to the observed expression levels of PtTIF5α. Amplification conditions were (1) incubation at 95°C for 1 min; then (2) 45 cycles of cDNA amplification at 95°C for 15 s, a specific annealing temperature for 20 s, and 72°C for 20 s (PtNASs, PtYSLs, PtFROs, PtFIT, Potri.006G074900, Potri.018G141800, and Potri.006G248500) or 30 s (PtTIF5α and PtIRTs). Each sample was analyzed as three technical replicates. The primer sequences and annealing temperatures used for real-time RT-PCR are shown in Supplemental Table 2.
2.5. Statistical analysis
For soil cultivation, three plants were grown on each soil type (normal soil or calcareous soil) to provide three biological replicates (n = 3). For hydroponic cultivation, four plants were grown on Fe-sufficient or Fe-deficient culture, providing four biological replicates (n = 4). For gene expression analysis, we selected two plants from the Fe-sufficient or Fe-deficient hydroponic culture for two biological replicates (n = 2). RNA was extracted from both plants, and three real-time RT-PCR reactions were performed for each RNA sample as technical replicates (n = 3). For plant height, main trunk diameter, and SPAD value data, we used the t-test with Microsoft Excel 2010 software. Then, analysis of variance was used with Student’s t-test to examine experimental SPAD value data, metal concentrations, and gene expression; we employed JMP10 software (SAS Institute, Cary, NC, USA) for these analyses and considered p < 0.05 statistically significant.
3. Results
3.1. Growth analysis for soil cultivation
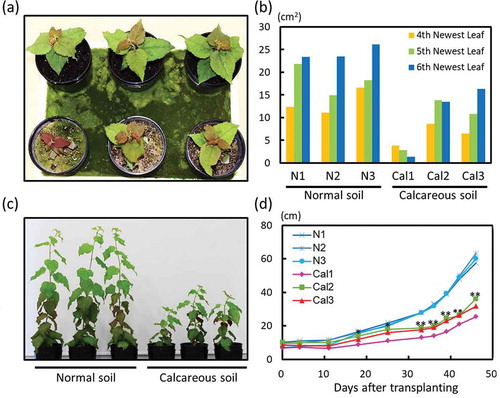
A growth experiment was performed on normal soil and calcareous soil. Although poplars did grow, they exhibited clear growth defects in calcareous soil (). The total leaf area of the fourth, fifth, and sixth newest leaves was smaller in plants grown on calcareous soil compared to those grown on normal soil ((,)). Plant growth differed markedly between the two soil types when plants were grown for 43 days (,)). However, the SPAD values of the new leaves of plants grown on normal and calcareous soils did not differ (Fig. S1).
Figure 1. Growth, symptoms, and total leaf area of poplar plants grown on normal or calcareous soil. (a) Plant appearance and growth 18 days after transplanting. Upper three plants: plants grown on normal soil; lower three plants: plants grown on calcareous soil. (b) Estimated sizes of individual leaves (fourth, fifth, and sixth newest leaves) 18 days after transplanting. Leaf size was estimated as length × width × 1/2 cm2. (c) Plant appearance and growth status 43 days after transplanting. (d) Plant height from 0 to 45 days after transplanting. Cal1, Cal2, Cal3: independent plants grown on calcareous soil; N1, N2, N3: independent plants grown on normal soil. Asterisks at the top of the graph indicate significant differences (*P < 0.05; **P < 0.01) in plant height between normal soil and calcareous soil cultivation, as determined via Student’s t-test.
3.2. Growth analysis for hydroponic cultivation
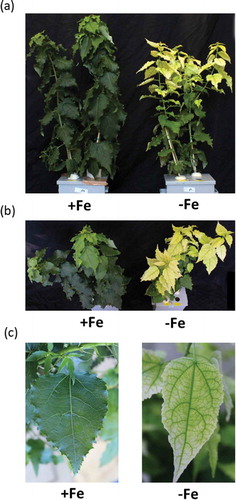
Poplars were cultivated in both Fe-sufficient and Fe-deficient hydroponic culture solutions twice (first and second cultivations). In the first hydroponic cultivation, Fe-deficient poplar was shorter than Fe-sufficient poplar after 18 days of Fe deficiency treatment (Fig. S2(a)). Then, poplar plants showed clear leaf chlorosis in new leaf after 27 days of Fe deficiency treatment (Fig. S2(b)). Chlorosis became increasingly serious in many leaves around 38–42 days of Fe deficiency (Fig. S2(c,d)). In the second hydroponic cultivation, poplars began to show chlorosis at 10 days of Fe deficiency, and root and leaf samples were taken for gene expression analysis at this stage (Fig. S3). At 35 days of Fe-deficiency treatment, very serious chlorosis was apparent in the new and middle leaves ((,)). The details of the normal green leaves of Fe-sufficient poplars and of the chlorotic leaves of 27-day Fe-deficient poplars are shown in ).
Figure 2. Growth and symptoms of poplar plants grown in hydroponic culture under Fe-sufficient or Fe-deficient conditions (second cultivation). Plant appearance and chlorosis symptoms front view (a) and top view (b) after 35 days of Fe-deficiency treatment. (c) Normal green leaf of Fe-sufficient plant and chlorotic leaf of Fe-deficient plant after 27 days of treatment.
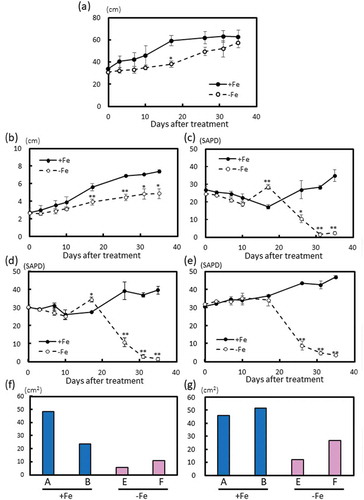
The plant height and the diameter of the main trunk were lower and smaller, respectively, in poplars under Fe-deficient conditions than in those with sufficient Fe ((,)). The SPAD values of the 5th, 7th, and 10th newest leaves clearly decreased between 16 and 34 days after beginning Fe-deficiency treatment ((–)). More severe chlorosis was apparent in the first to the fourth newest leaves, but these leaf sizes were too small to measure with the SPAD meter (data not shown). The leaf sizes of 35-day Fe-deficient poplars were smaller than those of Fe-sufficient poplars ((,)).
Figure 3. Plant growth, leaf SPAD value, and leaf area of Fe-sufficient or Fe-deficient poplar plants grown in hydroponic culture from 0 to 35 days after treatment (second cultivation). (a) Plant height (n = 4). (b) Diameters of the main trunk (n = 4). SPAD value of the 5th newest leaves (c), the 7th newest leaves (d), and the 10th newest leaves (e); (n = 4). Total leaf areas of the 10th newest leaves (f) and the 20th newest leaves (g) of Fe-sufficient or Fe-deficient poplar plants 35 days after treatment (n = 2). n represents biological replications. Asterisks above the bars indicate significant differences (*P < 0.05; **P < 0.01) between plants grown in Fe-sufficient and -deficient conditions. Alphabets (A, B, E, F) shown under graphs indicate plant ID of an individual poplar plant in second hydroponic cultivation.
The fluctuations of the pH in the Fe-sufficient and Fe-deficient hydroponic culture solutions are shown in Fig. S4. The pH value of the hydroponic culture solution was constant or increased in the Fe-sufficient solution. However, the pH value was constant or deceased in the Fe-deficient solution.
3.3. Metal concentrations in different leaves of poplars
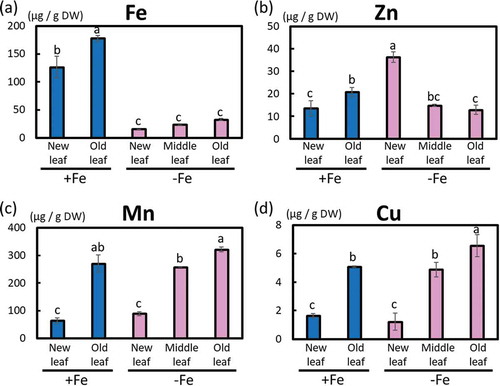
Fifty-four days after Fe-deficiency treatment, leaf SPAD values were measured and leaves from Fe-deficient and -sufficient cultivated poplars were sampled for Fe, Zn, Mn, and Cu concentration analysis (first hydroponic cultivation; Fig. S5). Our data show that the Fe concentration was obviously low in new leaves, middle leaves, and old leaves under Fe-deficient cultivation conditions ()). The Zn concentration was high in new leaves under Fe-deficient cultivation conditions ()). There was no clear difference in the Mn concentration in leaves between Fe-sufficient and -deficient cultivation conditions ()). Cu concentration was slightly higher in old leaves of Fe-deficient cultivation conditions ()).
Figure 4. Metal concentrations in different leaves of Fe-deficient and Fe-sufficient poplar plants grown in hydroponic culture 54 days after treatment (first cultivation). (a) Fe concentration, (b) Zn concentration, (c) Mn concentration, and (d) Cu concentration. Biological replication (n = 3). Error bar shows the standard error (SE). Values followed by different letters differed significantly according to Student’s t-test (P < 0.05).
3.4. Gene expression analysis in poplar roots and leaves
We analyzed gene expression levels in roots and leaves of Fe-sufficient and Fe-deficient poplars. RNA was extracted from 10-day Fe-sufficient or -deficient poplars from the second hydroponic cultivation (Fig. S3). Then, the relative expression levels of various Fe homeostasis-related genes were investigated using real-time RT-PCR (– and Fig. S7).
Figure 5. Relative expression levels of PtIRTs determined through real-time RT-PCR. (a) PtIRT1 and (b) PtIRT3 expressions in roots or fifth newest leaves of poplar plants grown in Fe-sufficient and Fe-deficient hydroponic cultures 10 days after treatment (second cultivation). Error bar shows the technical error, SE; n = 3. Data were normalized to the observed expression levels of PtTIF5α and displayed as relative gene expression (plant A, +Fe root = 1). Values followed by different letters differed significantly according to Student’s t-test (P < 0.05). Alphabets (A, B, C, D) shown under graphs indicate plant ID of an individual poplar plant in second hydroponic cultivation.
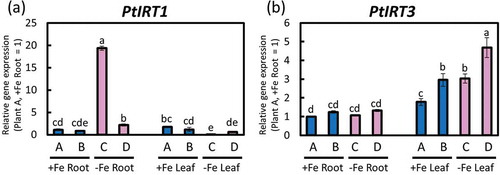
3.4.1. PtIRT expression in poplar roots and leaves
PtIRT1 was induced in roots under Fe deficiency as reported by Huang and Dai (Citation2015) ()). In our result, PtIRT3 might have been induced in Fe-deficient leaves, but there were no differences in the roots ()).
3.4.2. PtYSL expression in poplar roots and leaves
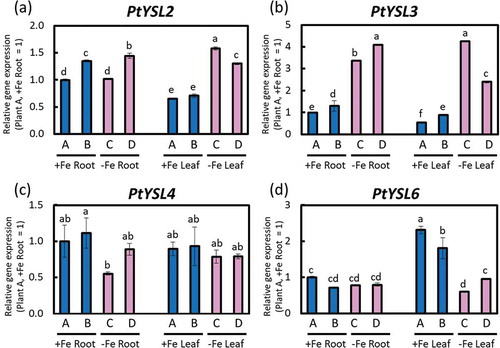
In this study, we referenced the report of Cao et al. (Citation2011) for poplar YSL genes. BLAST results of poplar YSL amino acid sequences are provided in the Supplemental Data 1, and a phylogenetic tree of PtYSLs is shown in Fig. S6. PtYSL2 was significantly induced in leaves under Fe deficiency ()). PtYSL3 was notably induced in both roots and leaves under Fe deficiency ()). In contrast, PtYSL4 expression might have been suppressed in Fe-deficient roots, and no expression changes were observed in leaves ()). PtYSL6 expression decreased obviously in leaves, but no change was observed in roots under Fe deficiency ()). PtYSL1 and PtYSL5 expression levels were also analyzed, but the clear gene expression results could not be obtained.
Figure 6. Relative expression levels of PtYSLs determined with real-time RT-PCR. (a) PtYSL2, (b) PtYSL3, (c) PtYSL4, and (d) PtYSL6 expression in roots or fifth newest leaves of Fe-sufficient and Fe-deficient poplar plants grown in hydroponic culture 10 days after treatment (second cultivation). Error bar shows the technical error, SE; n = 3. Data were normalized to the observed expression levels of PtTIF5α and presented as relative gene expression (plant A, +Fe root = 1). Values followed by different letters were significantly different according to Student’s t-test (P < 0.05). Alphabets (A, B, C, D) shown under graphs indicate plant ID of an individual poplar plant in second hydroponic cultivation.
3.4.3. PtNAS expression in poplar roots and leaves
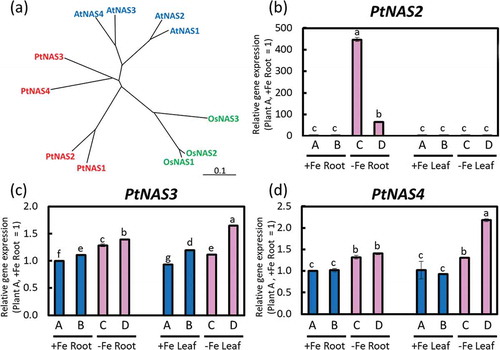
In this study, we defined four poplar NAS genes: PtNAS1, PtNAS2, PtNAS3, and PtNAS4. BLAST results of the amino acid sequences and a phylogenetic tree of PtNAS genes are shown in Supplemental Data 2 and ), respectively. There was high amino acid sequence similarity among Arabidopsis NASs, rice NASs, and poplar NASs. The phylogenetic tree shows that PtNAS forms an independent NAS group from AtNASs and OsNASs ()). PtNAS2 expression was significantly induced in Fe-deficient roots ()). PtNAS3 expression might be slightly elevated under Fe-deficient conditions ()). PtNAS4 might be induced in leaves under Fe-deficient conditions ()). Even though PtNAS1 expression level was also analyzed, however, the gene expression was not detected.
Figure 7. Relative expression levels of PtNASs based on real-time RT-PCR. (a) Phylogenetic tree of PtNASs. An unrooted phylogenetic tree of the NAS family from rice (OsNAS1‒3), Arabidopsis (AtNAS1‒4), and poplar (PtNAS1‒4). Homologues of PtNAS were searched and a phylogenetic tree was obtained using DNASIS Pro software (Hitachi Solutions, Ltd. Tokyo, Japan). Accession numbers of the amino acid sequences used to construct this phylogenetic tree are shown in Supplemental Table 1. (b) PtNAS2, (c) PtNAS3, and (d) PtNAS4 expression in roots or fifth newest leaves of Fe-sufficient and Fe-deficient poplar plants grown in hydroponic culture 10 days after treatment (second cultivation). Error bar shows the technical error, SE; n = 3. Data were normalized to the observed expression levels of PtTIF5α and displayed as relative gene expression (plant A, +Fe root = 1). Values followed by different letters differed significantly according to Student’s t-test (P < 0.05). Alphabets (A, B, C, D) shown under graphs indicate plant ID of an individual poplar plant in second hydroponic cultivation.
3.4.4. PtFRO expression in poplar roots and leaves
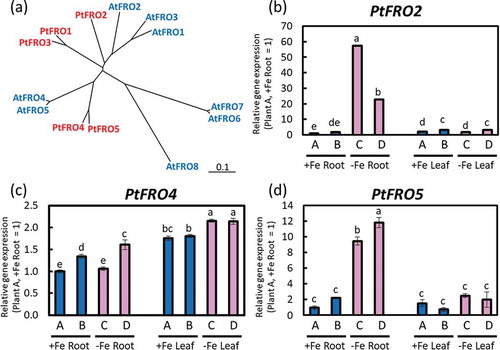
We defined five poplar FRO genes: PtFRO1, PtFRO2, PtFRO3, PtFRO4, and PtFRO5. BLAST results of the amino acid sequences and a phylogenetic tree are shown in Supplemental Data 3 and ), respectively. PtFRO2 had high homology with AtFRO2, which is induced in Fe-deficient roots (Robinson et al. Citation1999). Our results show that PtFRO2 and PtFRO5 were clearly induced in Fe-deficient roots (). PtFRO4 was slightly induced in leaves ()). Although PtFRO1 and PtFRO3 expression levels were analyzed, the gene expression was not detected.
Figure 8. Relative expression levels of PtFROs determined using real-time RT-PCR. (a) Phylogenetic tree of PtFROs. An unrooted phylogenetic tree of the FRO family from Arabidopsis (AtFRO1‒8) and poplar (PtFRO1‒5). Homologues of PtNAS were searched and a phylogenetic tree was obtained using DNASIS Pro software (Hitachi Solutions, Ltd. Tokyo, Japan). Accession numbers of the amino acid sequences used for this phylogenetic tree are provided in Supplemental Table 1. (b) PtFRO2, (c) PtFRO4, and (d) PtFRO5 expression in roots or fifth newest leaves of Fe-sufficient and Fe-deficient poplar plants grown in hydroponic culture 10 days after treatment (second cultivation). Error bar shows the technical error, SE; n = 3. Data were normalized to the observed expression levels of PtTIF5α and presented as relative gene expression (plant A, +Fe root = 1). Values followed by different letters differed significantly according to Student’s t-test (P < 0.05). Alphabets (A, B, C, D) shown under graphs indicate plant ID of an individual poplar plant in second hydroponic cultivation.
3.4.5. PtFIT expression in poplar roots and leaves
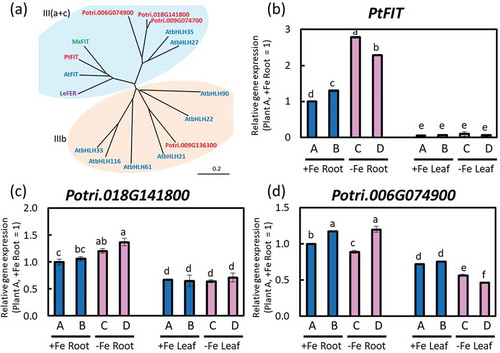
To find FIT genes in poplar, the amino acid sequence of AtFIT was used for BLAST. Poplar homologous genes were found as follows: Potri.009G005600 (66.98% similarity), Potri.009G136300 (34.36% similarity), Potri.006G074700 (53.55% similarity), Potri.018G141800 (54.14% similarity), and Potri.006G074900 (39.24% similarity). Phylogenic tree of AtbHLHs belonging to III (a + c) and III (b) subfamilies, MxFIT, LeFER, and poplar FIT homologous genes is shown in ). Only Pt009G005600 belongs to the FIT group. Thus, we defined Pt009G005600 as poplar FIT gene. There was high homology in the amino acid sequences of PtFIT, AtFIT, LeFER (tomato FIT: Ling et al. Citation2002) and MxFIT (apple genus FIT: Yin et al. Citation2014) (Supplemental Data 4). PtFIT was significantly induced in Fe-deficient roots ()). However, Potri.018G141800 and Potri.006G074900 were not induced under Fe deficiency ((c,d)). Although Potri.009G136300 and Potri.006G074700 expression levels were also analyzed, the gene expression was not detected.
Figure 9. Relative expression levels of PtFIT by real-time RT-PCR. (a) An unrooted phylogenetic tree of the bHLH III(a-c) and IIIb subfamily from Arabidopsis (AtFIT), poplar FIT homolog, LeFER and MxFIT. Phylogenetic tree was obtained using DNASIS Pro software (Hitachi Solutions, Ltd. Tokyo, Japan) based on amino acid sequences listed in Supplemental Table 1. (b) PtFIT, (c) Potri.018G141800, and (d) Potri.06G074900 expression in roots or fifth newest leaves of Fe-sufficient and Fe-deficient poplar plants grown in hydroponic culture 10 days after treatment (2nd cultivation). Error bar shows the technical error, SE; n = 3. Data were normalized to the observed expression levels of PtTIF5α and displayed as relative gene expression (plant A, +Fe root = 1). Values followed by different letters differed significantly according to Student’s t-test (P < 0.05). Alphabets (A, B, C, D) shown under graphs indicate plant ID of an individual poplar plant in second hydroponic cultivation.
3.4.6. Genes of the phenolic transporter, enzymes involved in coumarin biosynthetic pathway and H+-ATPase in poplar
To find the homologous genes of phenolic transporter in poplar, the amino acid sequence of AtABCG37/PDR9 (At3g53480) was used for BLAST. Poplar homologous genes were found as follows: Potri.001G189500 (45.92% similarity), Potri.002G188900 (74.90% similarity), Potri.002G189100 (75.45% similarity), Potri.006G248500 (70.32% similarity), Potri.008G099000 (77.88% similarity), Potri.010G153600 (76.94% similarity), Potri.010G153700 (76.73% similarity), Potri.010G153800 (77.15% similarity), and Potri.015G006000 (66.50% similarity). BLAST results of the amino acid sequences and a phylogenic tree of AtPDRs are shown in Supplemental Data 5 and Fig. S7(a), respectively. Then, we analyzed gene expression of Potri.006G248500 (AtPDR9 homolog) by real-time RT-PCR. Our results confirm that this gene was induced in Fe-deficient roots (Fig. S7(b)).
To find the homologous genes of enzymes involved in coumarin biosynthetic pathway in poplar, the amino acid sequence of F6′H1 (At3g13610), S8H (At3g12900), and CYP82C4 (At4g31940) was used for BLAST. Poplar homologous genes were found as follows: Potri.T170500 (83.93% similarity to F6′H1), Potri.001G006800 (83.15% similarity to F6′H1), Potri.001G007100 (80.06% similarity to F6′H1), Potri.001G006900 (83.90% similarity to F6′H1), Potri.005G097900 (81.77% similarity to S8H), Potri.014G037500 (83.63% similarity to CYP82C4), and Potri.014G037300 (84.02% similarity to CYP82C4). Amino acid sequences of the homologues of poplar F6′H1, S8H, and CYP82C4 are shown in Supplemental Data 6, 7, and 8, respectively.
To find the homologous genes of H+-ATPase involved in Fe uptake in poplar, BLAST search using the amino acid sequence of AHA2 (At4g30190) was also performed. Poplar homologous genes were found as follows: Potri.006G005900 (90.96% similarity), Potri.006G165900 (94.69% similarity), Potri.006G188600 (91.64% similarity), Potri.006G275000 (93.68% similarity), Potri.018G090300 (94.92% similarity), and Potri.018G112400 (91.30% similarity). The phylogenic trees or amino acid sequences of AHAs and the homologues of poplar are shown in Fig. S8 and Supplemental Data 9, respectively.
4. Discussion
4.1. Iron-deficiency symptoms exhibited by poplars grown in calcareous soil and hydroponic culture
Plants grown on calcareous soil with high pH often exhibit Fe-deficiency symptoms, leading to reduced crop yield and quality. Thus, Fe is a key determinant of biomass production and plant product quality (Briat et al. Citation2015). Poplar growth defects were obvious in calcareous soil cultivation and Fe-deficient hydroponic cultivation (–). The SPAD value of the new leaves clearly dropped during Fe-deficient hydroponic cultivation ((c–e)). However, the SPAD value of new leaves did not drop notably (Fig. S1), and Fe-deficiency chlorosis was not apparent seriously in poplar plants grown in calcareous soil compared to those grown in normal soil (). On the other hand, SPAD values of the new leaves of rice drop markedly, and rice plants show very clear Fe-deficiency chlorosis within 10 days under cultivation in calcareous soil (Masuda et al. Citation2017). Sweet potato, ravenna grass, and tobacco plants also exhibit Fe-deficiency chlorosis under calcareous soil cultivation (Nozoye et al. Citation2017a; Nozoye et al. Citation2017b; Xiong et al. Citation2014.). Thus, compared to rice, sweet potato, ravenna grass, or tobacco, poplars may have some tolerance for Fe deficiency when grown in calcareous soil.
4.2. Poplars may have excellent translocation ability from old leaves to new leaves
Poplars were able to survive under long-term Fe-deficiency treatment and did not die even after 61 days of Fe deficiency in hydroponic culture (Fig. S2(e)). Growth was affected, but poplars still grew under long-term Fe-deficiency conditions. According to our results, the Fe concentration was also lower in the old leaves of 54-day Fe-deficient-treated poplars ()). These old leaves existed since the plant was in Fe-sufficient conditions, and the SPAD value of these leaves was still high (Fig. S5). The difference in the Fe concentrations of the old leaves of Fe-sufficient and -deficient poplars shows that Fe might be translocated from old leaves to new leaves under Fe-deficient conditions.
Fe was not remobilized well under Fe-sufficient conditions. Maillard et al. (Citation2015) reported nutrient remobilization in leaves of various crops and tree species grown under normal field conditions or in soil under greenhouse conditions. In their report, Fe transport from old leaves was restricted in various crops and tree species during leaf senescence. Populus nigra L. did not exhibit Fe remobilization in leaves during field cultivation. However, we found that poplars may have a strong ability to translocate Fe from old leaves to new leaves when grown under Fe-deficient conditions. This may be the reason that poplars could grow and survive under long-term Fe-deficient conditions. Poplars may use Fe stored within the plant body efficiently. The Zn concentration increased in new leaves and decreased in old leaves under Fe-deficient conditions ()), which might have been due to the higher expression of Fe/Zn co-transporter genes, such as YSLs, or the higher concentrations of Fe/Zn chelators, such as NA. Our results indicate that PtYSL2, PtYSL3 and PtNAS4 expression increased in Fe-deficient leaves (, )).
After the resupply of Fe into the hydroponic solution, 61-day Fe-deficient poplar plants rapidly recovered, and the SPAD value of fifth newest leaves increased to more than 20 within 4 days and to more than 30 within 10 days (Fig. S2(e), data not shown). Thus, poplars may have a very robust ability for Fe remobilization and tolerance for Fe deficiency.
4.3. Poplar roots may interact with the rhizosphere to uptake Fe under Fe-deficient conditions
The nitrogen resource in the hydroponic culture solution used in our study was NO3 base (5 mM KNO3, 5 mM Ca(NO3)2 and 1 mM NH4NO3; see ‘Materials and methods’). Thus, whenever poplar plants consumed nitrogen from the hydroponic solution during cultivation, the pH of the solution should be gradually increased. In our experiment, the pH of the Fe-sufficient hydroponic solution increased during cultivation, but the pH of the Fe-deficient hydroponic solution was unchanged or even decreased (Fig. S4). Proton extrusion is induced by H+-ATPase under Fe deficiency in Arabidopsis roots (Santi and Schmidt Citation2009). We found that poplar also has AHA2 homologue (Fig. S8, Supplemental Data 9). Thus, poplar roots may also have the ability to decrease solution pH to change Fe in the rhizosphere into a soluble form and enhance Fe uptake by roots under Fe deficiency condition.
The color of the Fe-deficient hydroponic solution was yellower than the Fe-sufficient hydroponic solution (data not shown). There have been reports that Arabidopsis secretes phenolics, such as coumarins, from its roots by AtPDR9 (AtABCG37) to mobilize and take up Fe through the expression of AtFRO2 and AtIRT1 (Fourcroy et al. Citation2016). Rice also secretes phenolics (protocatechuic acid) from its roots to the rhizosphere under Fe deficiency (Ishimaru et al. Citation2011). Poplar also has Arabidopsis AtPDR9 homologue (Fig. S7(a), Supplemental Data 5). Among the homologous genes, Pori.006G248500 was induced in Fe-deficient roots (Fig. S7(b)), suggesting that this gene may work to export phenolics. Additionally, poplar also has homologous genes of enzymes involving the biosynthetic pathway of coumarins, F6′H1, S8H, and CYP82C4 (Supplemental Data 6–8, Rajniak et al. 2018). Thus, these results suggest that poplars might also release phenolics of some sort to enhance uptake Fe from the rhizosphere.
4.4. Iron-deficiency-inducible genes in poplars
We confirmed that PtIRT1 expression increased in Fe-deficient roots, as reported by Huang and Dai (Citation2015) ()). PtIRT1 expression was higher in Fe-deficiency poplar C (plant ID) than D perhaps because Fe deficiency was serious in poplar C than D. PtIRT3 was also induced in Fe-deficient leaves ()), suggesting that PtIRT3 might play a role in Fe transport from vasculature to leaf tissue in Fe-deficient poplars.
DiDonato et al. (Citation2004) suggested that the major function of AtYSL2 appears to be the lateral movement of metals in the vasculature. Our results show that PtYSL2 and PtYSL3 gene expression increased in Fe-deficient leaves and/or roots (), suggesting that poplar YSL may play a similar role in the lateral movement of Fe in the vasculature and translocation of Fe from roots to new leaves.
PtNAS2 was remarkably induced in Fe-deficient roots ()). Higher production of NA increases Fe concentration in rice seeds, and NA is thus a very effective enhancer of Fe translocation (Masuda et al. Citation2009; Lee et al. Citation2009). Schuler et al. (Citation2012) proposed that NA facilitates the transport of Fe from the phloem to sink organs in Arabidopsis. Thus, the NA produced in roots by PtNAS2 might chelate Fe storage in root tissues and enhance translocation from roots to sink organs, such as leaves, via phloem. PtNAS4 expression increased in Fe-deficient leaves ()). NA produced in leaves by PtNAS4 might support Fe remobilization in leaves or Fe movement from vasculature to leaf cells together with PtYSL2 and PtYSL3. Tomato mutant chloronerva lacks NA because the activity of only one NAS in tomato is lost (Higuchi et al. Citation1995) by the mutation in NAS (Ling et al. Citation1999). The mutant harbors a single base change compared with the wild type, and the phenylalanine (279th amino acid in sequence of Supplemental Data 2) is changed to serine. They concluded that this phenylalanine is essential for NAS activity. This phenylalanine is conserved in OsNAS1-3, HvNAS1, and AtNAS1-4, and they are functional (Inoue et al. Citation2003; Higuchi et al. Citation1999; Suzuki et al. Citation1999; Ushio et al. Citation2003). Similarly, we found that phenylalanine is also conserved in PtNAS2, 3, 4 (Supplemental Data 2), suggesting that these PtNASs are also functional. However, PtNAS1 lacks 18 amino acids from 276th to 295th including this 279th phenylalanine (Supplemental Data 2). Thus, it suggests that PtNAS1 may have no NAS activity similar to chloronerva NAS.
PtFRO2 and PtFRO5 were highly induced in Fe-deficient roots (). PtFRO2 is highly homologous (76% amino acid sequence similarity) to AtFRO2 (Supplemental Data 3). AtFRO2 is induced in Fe-deficient roots and contributes Fe reduction and Fe uptake in Arabidopsis (Robinson et al. Citation1999). Therefore, PtFRO2 and PtFRO5 may also be involved in reducing ferric-chelate to ferrous and contribute to the ferrous ion uptake of roots by PtIRT1 under Fe deficiency.
PtFIT was induced in Fe-deficient roots ()). The amino acid sequence of PtFIT has a 67.0% similarity to that of AtFIT (Supplemental Data 4). There was also a high similarity between PtFIT and LeFER or MxFIT. Thus, PtFIT might act as a transcription factor regulating Fe homeostasis-related genes in poplars.
We also analyzed the gene expression levels of PtYSL1, PtYSL5, PtNAS1, PtFRO1, PtFRO3, Potri.009G136300, and Potri.006G074700. However, the gene expression was not detected (data not shown). It is perhaps because the expression levels of these genes were too low in roots and leaves for detection with real-time RT-PCR.
4.5. Poplars are a highly suitable breeding target for improving Fe-deficiency tolerance in problem soils
There have been reports that plants with Fe-deficiency tolerance were obtained through gene-introduction approaches. Xiong et al. (Citation2014) reported that overexpression of AhIRT1 enhanced Fe-deficiency tolerance in tobacco plants. Oki et al. (Citation2004) artificially reconstructed and mutagenized the yeast Fe (III)-chelate reductase gene FRE1 to generate refre1/372, which encodes a protein that exhibits enhanced enzymatic activity at high pH to facilitate growth in calcareous soils. Then, Ishimaru et al. (Citation2007) and Masuda et al. (Citation2017) introduced refre1/372 into rice plants under the control of the OsIRT1 promoter to enhance the Fe-deficiency tolerance of rice plants. Hybrid poplar (P. tremula × P. tremuloides, T89) used in this research is available for transgenic approaches (Eriksson et al. Citation2000). Thus, the introduction of PtIRT1 promoter-driven refre1/372, leading to higher expression of refre1/372 in Fe deficiency root, has the potential to produce Fe-deficiency-tolerant poplars.
Drought-tolerance mechanisms in poplars have been reported (Regier et al. Citation2009; Khan et al. Citation2016). Drought-resistant poplars were generated by the introduction of the CpFATB gene or the Arabidopsis EDT1/HDG11 genes (Zhang et al. Citation2013; Yu et al. Citation2016). Takabe et al. (Citation2008) successfully produced a heat-resistant poplar through the introduction of the DnaK gene. Additionally, Sakamoto et al. (Citation2016) enhanced the wood-biomass production of poplar by increasing the expression of the rice secondary wall NAC domain protein 1, OsSWN1. Combining these approaches with Fe-deficiency tolerance will produce new poplars for desert greening and biomass production.
5. Conclusions
The overall results of this study show that poplars exhibit growth defects under Fe-deficient conditions. However, poplars may avoid Fe deficiency using mechanisms, such as adjusting the pH of the rhizosphere and translocating Fe from old leaves to new leaves under Fe-deficient conditions. We report Fe-deficiency-inducible genes in poplars, including PtYSL2, PtYSL3, PtNAS2, PtNAS4, PtFRO2, PtFRO5, and PtFIT. These genes may be involved in Fe uptake and in the translocation mechanisms in poplars under Fe-deficient conditions. Our findings may lead to a better understanding of the Fe-deficiency response in poplars and the breeding of Fe-deficiency-tolerant poplars to increase biomass production, enhance the greening of high-pH soils, and overcome global warming.
Supplemental Material
Download Zip (2.8 MB)Acknowledgments
We thank the following individuals from Ishikawa Prefectural University: Dr. Tatsuro Hamada for research facility support and Ms. Rieko Umeno, Ms. Yukiko Sato, and Ms. Aki Okubo for research assistance. We also thank Ms. Shiho Kamikabeya (Forest Bio-Research Center, Forestry and Forest Products Research Institute) for research assistance. We are grateful to Nihonkai Kougyou, Toyama, Japan, for kindly providing calcareous soil.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary data can be accessed here.
Additional information
Funding
References
- Adler PR, Grosso SJD, Parton WJ 2007: Life-cycle assessment of net greenhouse-gas flux for bioenergy cropping systems. Ecological Appli., 17, 675–691. doi: 10.1890/05-2018
- Briat JF, Dubos C, Gaymard F 2015: Iron nutrition, biomass production, and plant product quality. Trends Plant Sci., 20, 33–40. doi: 10.1016/j.tplants.2014.07.005
- Brunner AM, Yakovlev IA, Strauss SH 2004: Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol., 4, 14. doi: 10.1186/1471-2229-4-14
- Cao J, Huang J, Yang Y, Hu X 2011: Analyses of the oligopeptide transporter gene family in poplar and grape. BMC Genomics., 12, 465. doi: 10.1186/1471-2164-12-465
- Colangelo EP, Guerinot ML 2004: The essential Basic Helix-Loop-Helix protein FIT1 is required for the iron deficiency response. Plant Cell., 16, 3400–3412. doi: 10.1105/tpc.104.024315
- Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML 2003: Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol., 133, 1102–1110. doi: 10.1104/pp.103.025122
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL 2001: Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature, 409, 346–349. doi: 10.1038/35053080
- DiDonato RJ, Roberts LA, Sanderson T, Eisley RB, Walker EL 2004: Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine–metal complexes. Plant J., 39, 403–414. doi: 10.1111/j.1365-313X.2004.02128.x
- Eide D, Broderius M, Fett J, Guerinot ML 1996: A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA., 93, 5624–5628. doi: 10.1073/pnas.93.11.5624
- Eriksson ME, Israelsson M, Olsson O, Moritz T 2000: Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol., 18, 784–788. doi: 10.1038/77355
- FAO-GIS 1998: Distribution of Calcisols http://www.fao.org/soils-portal/soil-management/management-of-some-problem-soils/calcareous-soils/en/
- Fourcroy P, Sisó-Terraza P, Sudre D, Savirón M, Reyt G, Gaymard F, Abadía A, Abadia J, Álvarez-Fernández A, Briat JF 2014: Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New. Phytologist., 201, 155–167. doi: 10.1111/nph.12471
- Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C 2016: Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe2+ transport system. Mol. Plant., 9, 485–488. doi: 10.1016/j.molp.2015.09.010
- Gordon JC 2001: Poplars: trees of the people, trees of the future. For. Chron., 77, 217–219. doi: 10.5558/tfc77217-2
- Hell R, Stephan UW 2003: Iron uptake, trafficking and homeostasis in plants. Planta, 216, 541–551.
- Higuchi K, Kanazawa K, Nishizawa NK, Chino M, Mori S 1994: Purification and characterization of nicotianamine synthase from Fe- deficient barley roots. Plant Soil., 165, 173–179. doi: 10.1007/BF00008059
- Higuchi K, Nishizawa NK, Yamaguchi H, Roemheld V, Marschner H, Mori S 1995: Response of nicotianamine synthase activity to Fe deficiency in tobacco plants as compared with barley. J. Exp. Bot., 46, 1061–1063. doi: 10.1093/jxb/46.8.1061
- Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S 1999: Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol., 119, 471–479. doi: 10.1104/pp.119.2.471
- Huang D, Dai W 2015: Two iron-regulated transporter (IRT) genes showed differential expression in poplar trees under iron or zinc deficiency. J. Plant Physiol., 186–187, 59–67. doi: 10.1016/j.jplph.2015.09.001
- IGBP-DIS 1998: SoilData(V.0) A Program for Creating Global Soil-Property Databases. IGBP Global Soils Data Task, France.
- Inoue H, Higuchi K, Takahashi M, Nakanishi H, Mori S, Nishizawa NK 2003: Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long‐distance transport of iron and differentially regulated by iron. Plant J., 36, 366–381. doi: 10.1046/j.1365-313X.2003.01878.x
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK 2009: Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem., 284, 3470–3479. doi: 10.1074/jbc.M806042200
- Isebrands JG, Sturos JA, Crist JB 1979: Integrated utilization of biomass, a short-rotation intensively cultured populous raw material. TAPPI, 627, 67–70.
- Ishimaru Y, Kakei Y, Shimo H, Bashir K, Sato Y, Sato Y, Uozumi N, Nakanishi H, Nishizawa NK 2011: A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J. Biol. Chem., 286, 24649–24655. doi: 10.1074/jbcM111.221168
- Ishimaru Y, Kim S, Tsukamoto T et al. 2007: Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc. Nat. Aca. Sci. USA., 104, 7373–7378. doi: 10.1073/pnas.0610555104
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P 2004: FRU (BHLH029) is required for induction of iron mobilization genes. In Arabidopsis Thaliana. FEBS Lett, 577., 528–534.
- Jean ML, Schikora A, Mari S, Briat JF, Curie C 2005: A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J., 44, 769–782. doi: 10.1111/j.1365-313X.2005.02569.x
- Keller T, Koch W 1964: The effect of iron chelate fertilization of poplar upon CO2-uptake, leaf size, and content of leaf pigments and iron. Plant Soil., 20, 116–126. doi: 10.1007/BF01378103
- Khan Z, Rho H, Firrincieli A, Hung SH, Luna V, Masciarelli O, Kim SH, Doty SL 2016: Growth enhancement and drought tolerance of hybrid poplar upon inoculation with endophyte consortia. Curr. Plant., 6, 38–47. doi: 10.1016/j.cpb.2016.08.001
- Kobayashi T, Shinkai A, Yasufuku N, Omine K, Marui A, Nagafuchi T 2012: Field surveys of soil conditions in steppe of northeastern mongolia. J. Arid Land Studies, 22, 25–28.
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK 2004: OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J., 39, 415–424. doi: 10.1111/j.1365-313X.2004.02146.x
- Lee S, Jeon US, Lee SJ et al. 2009: Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. USA, 106, 22014–22019. doi: 10.1073/pnas.0910950106
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M 2002: The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. USA, 99, 13938–13943. doi: 10.1073/pnas.212448699
- Ling HQ, Koch G, Bäumlein H, Ganal MW 1999: Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc. Natl. Acad. Sci. USA, 96, 7098–7103. doi: 10.1073/pnas.96.12.7098
- Maillard A, Diquélou S, Billard V, Laîné P, Garnica M, Prudent M, Garcia-Mina JM, Yvin JC, Ourry A 2015: Leaf mineral nutrient remobilization during leaf senescence and modulation by nutrient deficiency. Front. Plant Sci., 6, 317. doi: 10.3389/fpls.2015.00317
- Marschner H 1995: Mineral Nutrition of Higher Plants. 2nd. Academic Press, London.
- Masuda H, Shimochi E, Hamada T, Senoura T, Kobayashi T, Aung MS, Ishimaru Y, Ogo Y, Nakanishi H, Nishizawa NK 2017: A new transgenic rice line exhibiting enhanced ferric iron reduction and phytosiderophore production confers tolerance to low iron availability in calcareous soil. PLoS ONE, 12, e0173441. doi: 10.1371/iournal.pone.0173441
- Masuda H, Usuda K, Kobayashi T, Ishimaru Y, Kakei Y, Takahashi M, Higuchi K, Nakanishi H, Mori S, Nishizawa NK 2009: Overexpression of the barley nicotianamine synthase gene HvNAS1 increase iron and zinc concentrations in rice grains. Rice, 2, 155–166. doi: 10.1007/s12284-009-9031-1
- Migeon A, Blaudez D, Wilkins O, Montanini B, Campbell MM, Richaud P, Thomine S, Chalot M 2010: Genome-wide analysis of plant metal transporters, with an emphasis on poplar. Cell. Mol. Life Sci., 67, 3763–3784. doi: 10.1007/s00018-010-0445-0
- Nozoye T, Aung MS, Masuda H, Nakanishi H, Nishizawa NK 2017b: Bioenergy grass [Erianthus ravennae (L.) Beauv.] secretes two members of mugineic acid family phytosiderophores which involved in their tolerance to Fe deficiency. Soil. Sci. Plant Nutri., 63, 543–552. doi: 10.1080/00380768.2017.1394168
- Nozoye T, Otani M, Senoura T, Nakanishi H, Nishizawa NK 2017a: Overexpression of barley nicotianamine synthase 1 confers tolerance in the sweet potato to iron deficiency in calcareous soil. Plant Soil., 418, 75–88. doi: 10.1007/s11104-016-3134-4
- Oki H, Kim S, Nakanishi H, Takahashi M, Yamaguchi H, Mori S, Nishizawa NK 2004: Directed evolution of yeast ferric reductase to produce plants with tolerance to iron deficiency in alkaline soils. Soil Sci. Plant Nut ., 50, 1159–1165. doi: 10.1080/00380768.2004.10408589
- Pich A, Scholz G 1996: Translocation of copper and other micronutrients in tomato plants (Lycopersicon esculentum Mill.): nicotianamine-stimulated copper transport in the xylem. J. Exp. Bot., 294, 41–47. doi: 10.1093/jxb/47.1.41
- Pires N, Dolan L 2010: Origin and diversification of basic-Helix-Loop-Helix proteins in plants. Mol. Biol. Evol., 27, :862–874. doi: 10.1093/molbev/msp288
- Rajniak J, Giehl RFH, Chang E, Murgia I, Von Wirén N, Sattely ES 2018: Biosynthesis of redox-active metabolites in response to iron deficiency in plants. Nat. Chem. Biol., 14, 442–450. doi: 10.1038/s41589-018-0019-2
- Ramon Magsaysay award of Peace and International Understanding, 2003 http://rmaward.asia/awardees/toyama-seiei/
- Regier N, Streb S, Cocozza C, Schaub M, Cherubini P, Zeeman SC, Frey B 2009: Droughttolerance of two black poplar (Populus nigra L.) clones: contribution of carbohydrates and oxidative stress defence. Plant Cell Environ., 32, 1724–1736. doi: 10.1111/j.1365-3040.2009.02030.x
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML 1999: A ferric-chelate reductase for iron uptake from soils. Nature, 397, 694–697. doi: 10.1038/17800
- Römheld V, Marschner M 1986: Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol., 80, 175–180. doi: 10.1104/pp.80.1.175
- Rudolph A, Becker R, Scholz G, Procházka Z, Toman J, Macek T, Herout V 1985: The occurrence of the amino acid nicotianamine in plants and microorganisms. A reinvestigation. Biochem. Physiol. Pflanzen., 180, 557–563. doi: 10.1016/S0015-3796(85)80036-6
- Sakai A, Larcher W 1987: Frost Survival of Plants: responses and Adaptation to Freezing Stress. Ecological Studies. 62. Springer-Verlag, Berlin Heidelberg Germany. ISBN 978-3-642-71745-1. doi: 10.1007/978-3-642-71745-1
- Sakamoto S, Takata N, Oshima Y, Yoshida K, Taniguchi T, Mitsuda N 2016: Wood reinforcement of poplar by rice NAC transcription factor. Sci. Rep., 6, 19925. doi: 10.1038/srep19925
- Santi S, Schmidt W 2009: Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New. Phytologist., 183, 1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x
- Schmid NB, Giehl RFH, Döll S, Mock HP, Strehmel N, Scheel D, Kong X, Hider RC, Von Wirén N 2014: Feruloyl-CoA 6ʹ-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol, 164, 160–172.
- Schuler M, Rellán-Álvarez R, Fink-Straube C, Abadía J, Bauer P 2012: Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell., 24, 2380–2400. doi: 10.1105/tpc.112.099077
- Sisó-Terraza P, Luis-Villarroya A, Fourcroy P, Briat J-F, Abadía A, Gaymard F, Abadía J, Álvarez-Fernández A 2016: Accumulation and secretion of coumarinolignans and other coumarins in Arabidopsis thaliana roots in response to iron deficiency at high pH. Front. Plant Sci., 7, 1711. doi: 10.3389/fpls.2016.01711
- Stanton B, Eaton J, Johnson J, Rice D, Schuette B, Moser B 2002: Hybrid poplar in the Pacific Northwest: the effects of market-driven management. J. For., 100, 28–33.
- Stephan UW, Schmidke I, Stephan VW, Scholz G 1996: The nicotianamine molecule is made-to-measure for complexation of metal micronutrients in plants. Biometals, 9, 84–90. doi: 10.1007/BF00188095
- Suzuki K, Higuchi K, Nakanishi H, Nishizawa NK, Mori S 1999: Cloning of nicotianamine synthase genes from Arabidopsis thaliana.. Soil Sci. Plant Nutr., 45, 993–1002. doi: 10.1080/00380768.1999.10414350
- Takabe T, Uchida A, Shinagawa F, Terada Y, Kajita H, Tanaka Y, Takabe T, Hayashi T, Kawai T, Takabe T 2008: Overexpression of DnaK from a halotolerant cyanobacterium Aphanothece halophytica enhances growth rate as well as abiotic stress tolerance of poplar plants. Plant Growth Regul., 56, 265–273. doi: 10.1007/s10725-008-9306-3
- Takahashi M, Terada Y, Nakai I, Nakanishi H, Yoshimura E, Mori S, Nishizawa NK 2003: Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell., 15, 1263–1280.
- Townsend RA, Kar SP, Miller RO 2014: Poplar (Populus spp.) Trees for Biofuel Production. eXtension 70456 http://articles.extension.org/pages/70456/poplar-populus-spp-trees-for-biofuel-production
- Tuskan GA, DiFazio S, Jansson S et al. 2006: The Genome of Black Cottonwood. Populus Trichocarpa (Torr. Gray). Science, 313, 1596–1604. doi: 10.1126/science.1128691
- Ushio Y, Mori S, Nishizawa NK 2003: Novel nicotianamine synthase and the AtNAS4 gene in Arabidopsis. Japanese Patent No. 2003–70926.
- Xiong H, Guo X, Kobayashi T et al. 2014: Expression of peanut Iron Regulated Transporter 1 in tobacco and rice plants confers improved iron nutrition. Plant Physiol. Biochem., 80, 83–89. doi: 10.1016/j.plaphy.2014.03.021
- Yin L, Wang Y, Yuan M, Zhang X, Xu X, Han Z 2014: Characterization of MxFIT, an iron deficiency induced transcriptional factor in Malus xiaojinensis. Plant Physiol. Biochem., 75, 89e95. doi: 10.1016/j.plaphy.2013.12.007
- Yu LH, Wu SJ, Peng YS et al. 2016: Arabidopsis EDT1/HDG11 improves drought and salt tolerance in cotton and poplar and increases cotton yield in the field. Plant Biotechnol. J., 14, 72–84. doi: 10.1111/pbi.12358
- Yuan YX, Zhang J, Wang DW, Ling HQ 2005: AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res., 15, 613–621. doi: 10.1038/sj.cr.7290331
- Zhang L, Liu M, Qiao G, Jiang J, Jiang Y, Zhuo R 2013: Transgenic poplar “NL895ʹ’ expressing CpFATB gene shows enhanced tolerance to drought stress. Acta. Physiol. Plant, 35, 603–613. doi: 10.1007/s11738-012-1101-0
