Abstract
AIMS: To explore and validate the utility of rumen endoscopy for collection of rumen papillae for gene expression measurement.
METHODS: Four adult Coopworth ewes were fasted for either 4 or 24 hours. Animals were sedated, placed in a dorsally recumbent position at 45 degrees with the head upright, and an endoscope inserted via a tube inserted into the mouth. Biopsies of rumen papillae were taken from the ventral surface of the rumen atrium under visual guidance. Two biopsies were collected from one of the animals that had been fasted for 4 hours, and three from one of the animals that had been fasted for 24 hours. Video of the rumen atrium and reticulum was also collected. The animals recovered uneventfully. Biopsies were subsequently used for extraction and sequencing of mRNA.
RESULTS: The ventral surface of the rumen atrium was accessible after 4 hours off pasture, but a larger region was accessible after 24 hours of fasting. Sedation allowed access for endoscope use for around 5 to 10 minutes after which increased saliva flow was noted. Rumen papillae biopsies were easily collected, with samples from a variety of sites collected in the ∼10 minute time window. High quality RNA was obtained for stranded mRNA sequencing. Of the resulting reads, 69–70% mapped uniquely to version 3.1 of the ovine genome, and 48–49% to a known gene. The rumen mRNA profiles were consistent with a previously reported study.
CONCLUSIONS: This method for obtaining rumenal tissue was found to be rapid and resulted in no apparent short or long term effects on the animal. High quality RNA was successfully extracted and amplified from the rumen papillae biopsies, indicating that this technique could be used for future gene expression studies. The use of rumen endoscopy could be extended to collection of a variety of rumen and reticulum anatomical measurements and deposition and retrieval of small sensors from the rumen. Rumen endoscopy offers an attractive and cost effective approach to repeated rumen biopsies compared with serial slaughter or use of cannulated animals.
Introduction
In recent years there has been increasing interest in understanding ruminant digestion, driven by the objective of genetically mitigating emissions of methane by ruminants (Clark Citation2013), either through direct selection on methane-yield (Pinares-Patiño et al.. Citation2013), or improved ruminant feed efficiency (Basarab et al. Citation2013). Improvement of these traits through genomic selection is currently being investigated (Pickering et al. Citation2015), and as a consequence the number of animals that have rumen samples being collected for microbial analysis has increased rapidly from 10–20 to thousands per annum. With the recent publication of repeatable and detailed differences in rumen contents between animals with differing methane-yield phenotypes (Kittelmann et al. Citation2014; Shi et al. Citation2014) interest has extended to the rapid sampling of rumen contents. Separately, the recent publication of the sheep genome and transcriptome demonstrated that high expression of genes encoding keratin cross-linking proteins was associated with adaptation to a ruminant lifestyle (Jiang et al. Citation2014).
Historically, rumen gene expression studies have involved either slaughter of animals (e.g. Naeem et al. Citation2012), or rumen cannulation, also referred to as fistulation (e.g. Steele et al. Citation2011). Both these methods are expensive, time consuming and not suitable for animals that are to be retained for breeding. Moreover, slaughter of the animal is not feasible for time-course studies as serial sampling of the same animals is impossible. Serial sampling can be undertaken using rumen cannulation, which allows long-term, minimally invasive access to the rumen (Hecker Citation1969), however, this process is not convenient for studies requiring large numbers of animals.
While cannulation is currently the conventional method for studies that require sequential sampling of rumen contents, endoscopic biopsies are a viable alternative. In domestic ruminants endoscopy via a ruminal cannula has been used for evaluation of the rumen (McBride et al. Citation1983), and for sampling to determine gene expression (Suominen et al. Citation1998; Taylor-Edwards et al. Citation2010; Steele et al. Citation2015). An alternative method of obtaining tissue for gene expression studies is via oral endoscopy. In calves, oral endoscopy has been evaluated as a tool to describe the visible structures of the rumen. This technique was found to be unsatisfactory in comparison to use of a ruminal cannula; however, the animals were not sedated during the procedure. As a result endoscopic examination of the rumen was only successful in 4/9 calves examined (Franz et al. Citation2006).
The objective of this study was to explore the viability of oral endoscopy for biopsy of rumen papillae, and evaluate the biopsies obtained via this method for RNA quality and gene expression analysis.
Materials and methods
Animals, sedation and restraint
All work was undertaken by approval of the AgResearch Invermay Animal Ethics Committee (Approval number: 13219).
Four adult female Coopworth sheep weighing 54–73 kg were fasted; two for 24 hours and two for 4 hours prior to sedation. They were each injected I/M with 2 mL atropine (0.65 mg/mL; Phoenix Atropine injection; Phoenix Pharm Distributors Ltd, Auckland, NZ), to reduce salivation and to stabilise the heart rate, followed approximately 10 minutes later by slow I/V xylazine (Xylase 20, Bayer Animal Health, Auckland, NZ) as detailed in . Each sheep was then placed in dorsal recumbency and lightly restrained in a modified laparoscopy cradle so that it could be positioned at 45 degrees for the procedure. This allowed the papillae in the anterio-ventral region of the rumen to be visualised in the gas dome of the rumen. The procedure was completed between 10 and 24 minutes after initial administration of xylazine () when the animal was removed from the cradle and returned to its pen. At this stage they could walk and stand, although initially mildly ataxic.
Table 1. Details of sheep using to assess oral endoscopy for biopsy of the rumen, with the dose of atropine and xylazine given to establish sedation, and the duration of the procedure.
Endoscopy
Following sedation and restraint in the modified laparoscopy cradle, a tube was introduced into the sheep's mouth and pharynx to protect the sheep and endoscope, and held by hand. The tube consisted of a PVC pipe (20 mm wide and 200 mm long), surrounded by a rubber dairy teat liner (DeLaval liner 24M; DeLaval Ltd, Hamilton, NZ) cut to fit over the pipe. A standard Olympus colonoscope (Olympus Flexible Video Evis CF-140L, Olympus, Auckland, NZ) with a diameter of 12.5 mm and a length of 170 cm connected to a light source and processor (Olympus Evis Exera II CV-180 and CLV-180) was introduced through the pipe and advanced through the oesophagus and into the rumen with visual guidance (Olympus OEV191H monitor). Following visual identification of the correct location of the papillae in the anterio-ventral region of the rumen, biopsies were taken using single-use biopsy forceps (Olympus Disposable Biopsy Forceps FB-230U) introduced through the instrumentation channel. Two rumen papillae biopsies were collected from one of the animals that had been fasted for 4 hours, and three biopsies from one of the animals that had been fasted for 24 hours. A video of the procedure and collection of biopsies is available (Supplementary Information 1Footnote1 and https://youtu.be/7CI0m9KPtUo). Between sheep, the outside of the instrument was cleaned with detergent (Medivators Intercept Detergent, CR Kennedy (NZ) Ltd., Auckland, NZ), the working channel was cleaned with a Pull-thru (CR Kennedy (NZ) Ltd.) to remove any remaining tissue and flushed with detergent. Rumen papillae biopsies were snap-frozen in liquid nitrogen then stored at –80 °C.
RNA extraction and sequencing
Total RNA was isolated from ∼10 mg of tissue using an RNeasy mini kit and an on-column DNase digestion (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA integrity was assessed using an Agilent RNA 6000 Nano Assay on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), and total RNA was quantified using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA).
The MiSeq Reagent Nano Kit v2 (Illumina, San Diego, CA, USA) was utilised for this proof-of-concept experiment. This kit is only capable of producing a maximum of one million reads, therefore, the two samples with the highest RNA integrity number (RIN; with a scale of 1–10, where 1 is completely degraded and 10 intact; Schroeder et al. Citation2006) were chosen for sequencing. Illumina TruSeq Stranded mRNA libraries (Illumina) were prepared according to the manufacturer's instructions. Libraries were visualised using an Agilent DNA High Sensitivity assay on an Agilent 2100 Bioanalyzer, and quantified using the Qubit dsDNA HS assay (Invitrogen, Life Technologies, Carlsbad CA, USA) according to the manufacturer's instructions. Qubit concentrations were used to pool the two indexed cDNA libraries containing the specific Illumina TruSeq adapters. The denatured cDNA libraries were pooled together with 1% PhiX control (Illumina), which acted as a quality control for cluster generation, sequencing, and alignment. Two pM of the combined sample library and PhiX control was loaded on an Illumina MiSeq (Illumina) for 300 cycles, resulting in paired-end reads of 150 base pairs. The Illumina adapters were removed from the sequences, along with bases with a median Phred scaled quality score below 20, using Trim Galore (version 0.3.7, Babraham Bioinformatics, Cambridge, UK). Trimmed reads were mapped to version 3.1 of the ovine reference genome (Jiang et al. Citation2014) using STAR (Dobin et al. Citation2013), with the supplied Ensembl Ovis aries transcriptome annotation (release 78). Only uniquely mapped reads with a maximum of two mismatches to the reference genome were retained for further analysis. The mapped reads were used to estimate raw read counts per gene using HTSeq (version 0.6.1p1, Anders et al. Citation2015) with the union overlap resolution mode.
Results
Biopsy using oral endoscopy
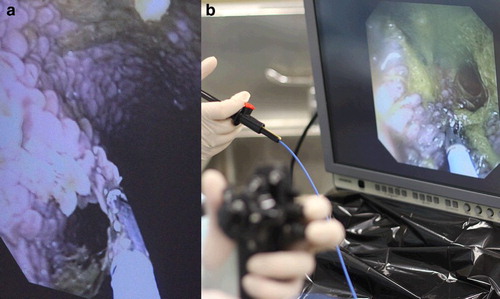
After the endoscope was passed into the rumen of the sheep the ventral surface of the rumen atrium was accessible after 4 hours off pasture, but a larger region was accessible after 24 hours of fasting. We were able to visualise the papillae in the anterio-ventral region of the rumen easily, but the remainder of the rumen was filled with debris in sheep fasted for either 4 hours or 24 hours. Some improvement was achieved by insufflating air into the lumen of the rumen. Sedation allowed access for endoscope use for around 5–10 minutes after which increased saliva flow was noted. Two to three biopsies were taken from each sheep under visual guidance (). The sheep tolerated the procedure without noticeable discomfort and recovered uneventfully. There was minimal bleeding following the sampling.
Figure 1. Photographs of (a) guided biopsy of papillae in the anterio-ventral region of the rumen of a sheep using oral endoscopy, and (b) the endoscopy procedure showing the hand held steering mechanism for the endoscope in the foreground, the hand piece to operate the biopsy forceps above this and the monitor showing the rumen with the papillae in the background.
RNA sequencing
Sample RIN for the five samples ranged from 6.3–8.2. The two samples with the highest RIN (≥8) were chosen for sequencing. They were both from the first sheep biopsied. Results for the two samples are presented in . Of the paired-end cDNA reads, 69–70% mapped to a unique region of the ovine genome, and 48–49% aligned to a known feature. The full list of read counts per gene is available in Supplementary Information 2Footnote2. The mRNA sequencing data have been deposited in the NCBI Gene Expression Omnibus Database, with the accession number GSE68791.
Table 2. Results for mRNA sequences obtained from two biopsy samples from a sheep rumen, acquired using endoscopy, showing the metrics generated by bioinformatics packages for alignment to the ovine genome (Jiang et al. Citation2014). The percentage of processed reads is given in brackets.
Discussion
Rumen biopsy using oral endoscopy was found to be rapid, able to undertaken in animals after 4 hours of fasting, and resulted in no apparent short or long term effects on the animal. Cannulation is currently the conventional method for studies that require sampling of rumen contents; however it is impracticable to repeat on a large scale. The alternative process of collecting samples post-slaughter cannot be used to collect samples serially. Perhaps most relevant to genetic studies, cannulated animals are very unlikely to be retained as part of an ongoing breeding flock or herd. Oral endoscopy, however, is a quick method to obtain rumen tissue biopsies that has no, or minimal, impact on the animal and can be done serially over time; while also imposing various treatments. A PVC pipe was used to protect not only the scope from the teeth of the sheep but also the sensitive oral surface from the scope.
While the integrity of the RNA obtained from the rumen biopsies varied, high quality RNA was able to be extracted for successful amplification. The quality of RNA may vary extensively between extractions (Schroeder et al. Citation2006), however steps can be taken to minimise degradation. These steps include reducing the time between sampling and immersing the tissue in liquid nitrogen, the addition of an RNA stabilisation reagent, and ensuring that the sample does not thaw before RNA extraction.
With optimal clustering, the kit that was used for this study is capable of producing one million reads, however our output was approximately 436,000 reads, under half of what was expected. This was the result of under-clustering of the flow cell due to the concentration of the library being under-estimated, and could be remedied by loading 6 pM of library onto the MiSeq instead of 2 pM. The gene expression profile was similar to previously reported rumen transcriptomes (Jiang et al. Citation2014). The two most highly expressed genes in both samples were the mitochondrial genes cytochrome c oxidase subunit I and cytochrome B.
The successful extraction and amplification of high quality RNA from the rumen biopsies indicates that this technique could be used for future gene expression studies. The amount of tissue could be increased if multiple biopsies were collected, or larger forceps used. Additionally, the use of an endoscope allows serial sampling of the same animal. Differences in the morphology (Bain et al. Citation2014), microbial community structure (Kittelmann et al. Citation2014) and microbiome (Shi et al. Citation2014) of the rumen have all been linked to methane-yield phenotypes in sheep. While these studies have primarily focussed on either the microbes or the host, in future the technique reported here could provide the opportunity to understand the interaction between the host transcriptome and the microbiome through serial sampling of both rumen papillae and rumen contents.
Although not investigated in the present study, rumen endoscopy could easily be extended to the collection of a variety of rumen and reticulum anatomical measurements and perhaps deposition and retrieval of small sensors from the rumen. There is precedence for this in humans, where pH and pressure sensors have been introduced via the nasogastric route for 24-hour measurements (Ghosh et al. Citation2011). A probe might also be clipped to the stomach/oesophageal wall for continuous wireless measurement (Chang et al. Citation2009).
In summary, biopsy via rumen endoscopy was found to be rapid, able to be undertaken in animals after 4 hours of fasting, and resulted in no apparent short or long term effects on the animal. High quality RNA was successfully extracted and amplified from the rumen papillae biopsies, indicating that this is a viable alternative for sample collection in future host gene expression studies. The use of rumen endoscopy could also easily be extended to collection of a variety of rumen and reticulum anatomical and pH measurements. Rumen endoscopy offers an attractive and cost effective approach to repeated rumen biopsies compared to serial slaughter or use of cannulated animals.
| RIN | = | RNA integrity number |
McRae_Supp_Info_1.mp4
Download MP4 Video (79.8 MB)McRae_Supp_Info_1.mp4
Download MP4 Video (79.8 MB)McRae_Supp._Info_2.pdf
Download PDF (7.7 MB)Acknowledgements
The authors wish to acknowledge Tracey van Stijn for her help in sequencing the libraries, Aaron Jeffs for supplying the mRNA library preparation kit reagents, Shannon Clarke for supplying the MiSeq Kit and Rudiger Brauning for his help with analysing the data. The equipment was provided by Gastroenterology Otago Ltd., Dunedin, New Zealand.
Notes
References
- Anders S, Pyl PT, Huber W. HTSeq — A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–69, 2015 doi: 10.1093/bioinformatics/btu638
- *Bain WE, Bezuidenhout L, Jopson NE, Pinares-Patiño CS, McEwan JC. Rumen differences between sheep identified as being low or high methane emitters. Proceedings of the 10th World Congress on Genetics Applied to Livestock Production 1–3. 2014
- Basarab JA, Beauchemin KA, Baron VS, Ominski KH, Guan LL, Miller SP, Crowley JJ. Reducing GHG emissions through genetic improvement for feed efficiency: effects on economically important traits and enteric methane production. Animal 7 (s2), 303–15, 2013 doi: 10.1017/S1751731113000888
- Chang JH, Choi MG, Yim DS, Cho YK, Park JM, Lee IS, Kim SW, Chung IS. A novel placement method of the Bravo wireless pH monitoring capsule for measuring intragastric pH. Digestive Diseases and Sciences 54, 578–85, 2009 doi: 10.1007/s10620-008-0399-3
- Clark H. Nutritional and host effects on methanogenesis in the grazing ruminant. Animal 7, 41–8, 2013 doi: 10.1017/S1751731112001875
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, 2013 doi: 10.1093/bioinformatics/bts635
- Franz S, Gentile A, Baumgartner W. Comparison of two ruminoscopy techniques in calves. The Veterinary Journal 172, 308–14, 2006 doi: 10.1016/j.tvjl.2005.06.008
- Ghosh T, Lewis DI, Axon ATR, Everett SM. Review article: methods of measuring gastric acid secretion. Alimentary Pharmacology and Therapeutics 33, 768–81, 2011. doi: 10.1111/j.1365-2036.2010.04573.x
- Hecker JF. A simple rapid method for inserting rumen cannulae in sheep. Australian Veterinary Journal 45, 293–4, 1969 doi: 10.1111/j.1751-0813.1969.tb01954.x
- Jiang Y, Xie M, Chen W, Talbot RT, Maddox JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 344, 1168–73, 2014 doi: 10.1126/science.1252806
- Kittelmann S, Pinares-Patiño CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC, Janssen PH. Two different bacterial community types are linked with the low-methane emission trait in sheep. Plos One 9, e103171, 2014 doi: 10.1371/journal.pone.0103171
- McBride BW, Berzins R, Milligan LP, Turner BV. Development of a technique for gastrointestinal endoscopy of domestic ruminants. Canadian Journal of Animal Science 63, 349–54, 1983 doi: 10.4141/cjas83-042
- Naeem A, Drackley JK, Stamey J, Loor JJ. Role of metabolic and cellular proliferation genes in ruminal development in response to enhanced plane of nutrition in neonatal Holstein calves. Journal of Dairy Science 95, 1807–20, 2012 doi: 10.3168/jds.2011-4709
- Pickering NK, Chagunda MGG, Banos G, Mrode R, McEwan JC, Wall E. Genetic parameters for predicted methane production and laser methane detector measurements. Journal of Animal Science 93, 11–20, 2015 doi: 10.2527/jas.2014-8302
- Pinares-Patiño CS, Hickey SM, Young EA, Dodds KG, MacLean S, Molano G, Sandoval E, Kjestrup H, Harland R, Hunt C, et al. Heritability estimates of methane emissions from sheep. Animal 7, 316–21, 2013 doi: 10.1017/S1751731113000864
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology 7, 3, 2006
- Shi W, Moon CD, Leahy SC, Kang D, Froula J, Kittelmann S, Fan C, Deutsch S, Gagic D, Seedorf H, et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Research 24, 1517–25, 2014 doi: 10.1101/gr.168245.113
- Steele MA, Dionissopoulos L, AlZahal O, Doelman J, McBride BW. Rumen epithelial adaptation to ruminal acidosis in lactating cattle involves the coordinated expression of insulin-like growth factor-binding proteins and a cholesterolgenic enzyme. Journal of Dairy Science 95, 318–27, 2011 doi: 10.3168/jds.2011-4465
- Steele MA, Schiestel C, AlZahal O, Dionissopoulos L, Laarman AH, Matthews JC, McBride BW. The periparturient period is associated with structural and transcriptomic adaptations of rumen papillae in dairy cattle. Journal of Dairy Science 98, 2583–95, 2015 doi: 10.3168/jds.2014-8640
- Suominen AH, Glimm DR, Tedesco D, Okine EK, McBurney MI, Kennelly JJ. Intestinal nutrient-gene interaction: the effect of feed deprivation and refeeding on cholecystokinin and proglucagon gene expression. Journal of Animal Science 76, 3104–13, 1998
- Taylor-Edwards CC, Burrin DG, Matthews JC, McLeod KR, Holst JJ, Harmon DL. Expression of mRNA for proglucagon and glucagon-like peptide-2 (GLP-2) receptor in the ruminant gastrointestinal tract and the influence of energy intake. Domestic Animal Endocrinology 39, 181–93, 2010 doi: 10.1016/j.domaniend.2010.05.002
