ABSTRACT
Aims
To apply molecular typing to DNA isolated from historical samples to determine Leptospira spp. infecting farmed and wild mammals in New Zealand.
Materials and methods
DNA samples used in this study were extracted from urine, serum or kidney samples (or Leptospira spp. cultures isolated from them) collected between 2007 and 2017 from a range of domestic and wildlife mammalian species as part of different research projects at Massey University. Samples were included in the study if they met one of three criteria: samples that tested positive with a lipL32 PCR for pathogenic Leptospira; samples that tested negative by lipL32 PCR but were recorded as positive to PCR for pathogenic Leptospira in the previous studies; or samples that were PCR-negative in all studies but were from animals with positive agglutination titres against serogroup Tarassovi. DNA samples were typed using PCR that targeted either the glmU or gyrB genetic loci. The resulting amplicons were sequenced and typed relative to reference sequences.
Results
We identified several associations between mammalian hosts and Leptospira strains/serovars that had not been previously reported in New Zealand. Leptospira borgpetersenii strain Pacifica was found in farmed red deer (Cervus elaphus) samples, L. borgpetersenii serovars Balcanica and Ballum were found in wild red deer samples, Leptospira interrogans serovar Copenhageni was found in stoats (Mustela erminea) and brushtail possums (Trichosurus vulpecula), and L. borgpetersenii was found in a ferret (Mustela putorius furo). Furthermore, we reconfirmed previously described associations including dairy cattle with L. interrogans serovars Copenhageni and Pomona and L. borgpetersenii serovars Ballum, Hardjo type bovis and strain Pacifica, sheep with L. interrogans serovar Pomona and L. borgpetersenii serovar Hardjo type bovis, brushtail possum with L. borgpetersenii serovar Balcanica, farmed deer with L. borgpetersenii serovar Hardjo type bovis and hedgehogs (Erinaceus europaeus) with L. borgpetersenii serovar Ballum.
Conclusions
This study provides an updated summary of host–Leptospira associations in New Zealand and highlights the importance of molecular typing. Furthermore, strain Pacifica, which was first identified as Tarassovi using serological methods in dairy cattle in 2016, has circulated in animal communities since at least 2007 but remained undetected as serology is unable to distinguish the different genotypes.
Clinical relevance
To date, leptospirosis in New Zealand has been diagnosed with serological typing, which is deficient in typing all strains in circulation. Molecular methods are necessary to accurately type strains of Leptospira spp. infecting mammals in New Zealand.
Introduction
Leptospirosis is an emerging zoonosis of global importance (Dunay et al. Citation2016). It is caused by the bacteria Leptospira, a genus with 71 species (Korba et al. Citation2021) and over 300 serovars (Picardeau Citation2017) worldwide. This genus is divided into two major clades: pathogenic (P) and saprophytic (S) and four subclades (P1, P2, S1 and S2). P1 is the virulent clade and P2 is the intermediate low-virulence clade, both with 21 species in each clade, while S1 and S2 are the saprophytic clades with 24 and 5 species in each clade, respectively (Vincent et al. Citation2019; Korba et al. Citation2021). The pathogenic species primarily infect mammals and can colonise a broad range of domestic and wild species (Picardeau Citation2017). Disease transmission to humans occurs via contact with infected animal urine or the contaminated environment. Historically in New Zealand, only two species from the P1 clade and six serovars were known to be endemic in animals (Marshall and Manktelow Citation2002). These included Leptospira borgpetersenii serovars Ballum, Tarassovi, Balcanica and Hardjo type bovis, and Leptospira interrogans serovars Pomona and Copenhageni. L. interrogans serovars Canicola and Australis have also been isolated but from human cases only (Marshall and Manktelow Citation2002), are observed occasionally in low numbers in humans, and are considered exotic (Nisa et al. Citation2020).
Surveillance of Leptospira strains circulating in New Zealand, both in humans and animals, relies on serology using the microscopic agglutination test (MAT) (Musso and La Scola Citation2013). However, typing strains with serology is challenging as this assay cannot distinguish between species, requires accurate knowledge of locally circulating strains, the interpretation of results can be subjective, and there is cross-reactivity within and between serogroups (Musso and La Scola Citation2013). For example, in New Zealand, cattle are often infected with serovar Hardjo type bovis which belongs to the species L. borgpetersenii. This strain serologically reacts like serovar Hardjo type prajitno which belongs to the species L. interrogans (Ramadass et al. Citation1990; Chideroli et al. Citation2017). Furthermore, both Hardjo type bovis and Hardjo type prajitno cross-react with serovar Balcanica as they are all in one serogroup, Sejroe. Serovars Hardjo type bovis and Balcanica are both endemic in New Zealand, but current typing methods using serology cannot distinguish between them. However, we recently developed a molecular typing method where we identified and demonstrated that genomic data from the glmU loci can be used to achieve serovar distinction for all serovars circulating in New Zealand, and it can discriminate all the pathogenic species in the P1 clade (Wilkinson et al. Citation2021).
In New Zealand, dairy farmers, meat workers and pig farmers were identified as high-risk occupations in the 1970s (Marshall and Manktelow Citation2002). Cattle were the recognised maintenance host for serovar Hardjo and pigs for serovars Pomona and Tarassovi (Marshall and Manktelow Citation2002). In the early 1980s, dairy cattle vaccines against serovars Hardjo and Pomona and pig vaccines against serovars Pomona and Tarassovi were implemented as an intervention to reduce the risk of disease in humans (Marshall and Chereshsky Citation1996; Fairly Citation1997). Analysis of serological data of notified human cases from 1999 to 2017 showed a decline in leptospirosis cases with serovars Hardjo (0.81–0.44 per 100,000) and Pomona (0.43–0.24 per 100,000) but an increase in cases with serovars Ballum (0.23–0.38 per 100,000) and Tarassovi (0.11–0.15 per 100,000) (Nisa et al. Citation2020). Tarassovi cases were associated with dairy farmers, while Ballum cases were associated with occupations other than farming and meat processing (Benschop et al. Citation2021). Serovar Ballum was first identified in the late 1970s in wildlife including the house mouse (Mus musculus), ship rat (Rattus rattus), brown rat (R. norvegicus), and hedgehog (Erinaceus europaeus) (Hathaway et al. Citation1978) and these host–serovar associations remain today (Moinet Citation2020). A study from 2016 to 2018 showed Ballum prevalence in mice varied seasonally from 83 (95% CI = 61–95)% to 86 (95% CI = 65–97)% in spring, to 31 (95% CI = 21–43)% to 37 (95% CI = 26–50)%, in autumn while prevalence in ship rats and hedgehogs was 44 (95% CI = 26–62)% and 27 (95% CI = 11–50)%, respectively (Moinet Citation2020). It is unclear whether the increase in human cases with serovar Ballum is linked to exposure to an increasing number of infected wild hosts, a more contaminated environment, or to a change in serovar-associations within livestock.
Serovar Tarassovi has historically been associated with pigs (Marshall and Manktelow Citation2002). However, a survey of 4,000 dairy cattle in 2016 identified 2.4% of cattle shedding Leptospira spp. and over 50% of the shedders were seropositive to Tarassovi. Since cattle have historically been associated with serovars Hardjo and Pomona, Tarassovi is now considered an emerging strain in dairy cattle posing public health risk (Yupiana et al. Citation2019a), especially for dairy farmers (Nisa et al. Citation2020). Application of molecular typing methods identified the strain shed by dairy cattle at the species level as L. borgpetersenii; however, the sequence was unlike serovar Tarassovi, thus this strain is called Pacifica (Bingham Citation2021; Wilkinson et al. Citation2021). Pacifica is currently included in serogroup Tarassovi since most animals shedding Pacifica have high titres against serovar Tarassovi, but no isolate has been obtained to date for confirmation by cross-agglutination absorption test (CAAT) (Babudieri Citation1971). It is unclear whether the increase in human cases of serovar Tarassovi is due to Tarassovi or Pacifica, as all human data to date has been acquired through serology and these two strains cannot be differentiated serologically.
Given the multi-host, multi-pathogen complexity of leptospirosis, the establishment of routine screening and typing as part of surveillance work in both wild and domesticated animals is necessary to fully understand its epidemiology. In this study, we used our recently developed molecular assay that is able to type all pathogenic species of Leptospira identified to date in the P1 clade and the serovars known to be endemic in New Zealand (Wilkinson et al. Citation2021) to identify Leptospira spp. in circulation in New Zealand mammals.
Materials and methods
Sample source
All DNA samples used in this study were collected as part of other research projects between 1 May 2007 and 18 November 2017 at the Molecular Epidemiology and Public Health Laboratory (mEpiLab; Massey University, Palmerston North, NZ) (Supplementary Table 1). Sample types included DNA extracted from serum, urine, and kidney, or cultures from these samples of domestic and wild mammals. A total of 997 DNA samples were tested in this study including samples from beef and dairy cattle (n = 774), farmed red deer (Cervus elaphus; n = 127), sheep (n = 25), wild red deer (Cervus elaphus; n = 24), brushtail possum (Trichosurus vulpecula; n = 30), European hedgehog (n = 7), stoat (Mustela erminea; n = 5), ship rat (n = 3), house mouse (n = 1) and ferret (Mustela putorius furo; n = 1) (). Domestic animal samples were collected from farms and abattoirs located in the lower North Island of New Zealand. Wildlife samples were collected via opportunistic sampling in and around Palmerston North. All DNA had been extracted using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instruction and stored at −20°C during the previous studies.
Table 1. Sample types from different mammals sourced between 2007 and 2017 in New Zealand, tested with lipL32, glmU and gyrB PCR to identify pathogenic Leptospira spp.
PCR
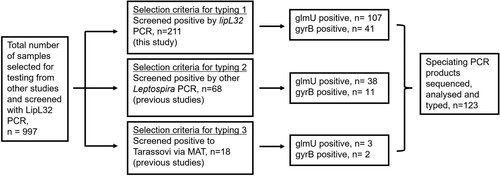
If PCR data were available from the previous studies, only samples that were previously positive for pathogenic Leptospira were tested in this study unless they met selection criteria for typing 3 (). This study used three different PCR assays as described previously (Wilkinson et al. Citation2021): a lipL32 quantitative PCR (qPCR) to screen for pathogenic Leptospira (Stoddard et al. Citation2009) which was used to identify Leptospira-positive samples, and a glmU (Wilkinson et al. Citation2021) or gyrB (Slack et al. Citation2006) speciating PCR which generated amplicons for sequencing to allow differentiation between different Leptospira taxa and their associated serovars. Samples were preferentially analysed with the glmU PCR as it can differentiate all seven endemic genotypes of Leptospira in New Zealand, while the gyrB locus has identical sequence for Balcanica and Tarassovi. If the glmU amplicons were insufficient in providing quality sequence data, then the samples were subjected to the gyrB PCR. Samples were selected for typing with the glmU or gyrB speciating PCR based on three selection criteria: (1) samples that tested positive with the lipL32 screening qPCR; (2) samples that tested negative to the lipL32 screening qPCR but were positive for PCR used to identify pathogenic Leptospira in previous studies; or (3) samples that were negative to PCR tests in all studies but were from animals with positive agglutination titres against serogroup Tarassovi in previous studies (). A cut-off of 96 was used to signify positive titres against Tarassovi, all of which were from dairy cattle.
Figure 1. Flowchart showing selection of DNA samples typed as part of a study of Leptospira spp. infection in mammals in New Zealand. Selection criteria for inclusion in the study were: criteria 1 – samples that were positive to lipL32 PCR; criteria 2 – samples negative to lipL32 PCR but positive to PCR for Leptospira spp. in previous studies; and criteria 3 – samples negative to lipL32 PCR but that came from animals seropositive to Tarassovi in previous studies. Samples were subjected to typing with the speciating PCR (glmU and/or gyrB) if they were positive to any one of these selection criteria.
A total of 45 amplification cycles were used for the lipL32 qPCR and a sample was considered positive if a quantification cycle (Cq) value was generated. The lipL32 qPCR assays were performed in sterile DNAse- and RNAse-free, 100-well rotor discs (Qiagen) in a Rotor-Gene 6000 PCR cycler (Corbett Research, Mortlake, Australia). Speciating PCR assays were performed using FIREPol master mix (Solis BioDyne, Tartu, Estonia) in sterile DNAse and RNAse-free 0.2-mL SnapStrip II PCR tubes (SSIbio, Lodi, CA, USA) in the Labcycler (SensoQuest, Göttingen, Germany) and visualised in an agarose gel. All primers used in this study are listed in Supplementary Table 2. All PCR assays included positive controls consisting of purified Leptospira DNA from laboratory reference strains of L. borgpetersenii serovar Hardjo type bovis and L. interrogans serovar Pomona. UltraPure distilled water (Invitrogen, Waltham, MA, USA) was used for no-template controls.
PCR purification and amplicon sequencing
The amplicons from the glmU and gyrB PCR were analysed by gel electrophoresis in 1% agarose gels stained with RedSafe (iNtRON Biotechnology, Seongnam, South Korea) and analysed under a Gel Doc XR UV transilluminator (Bio-Rad, Hercules, CA, USA). The gel bands of the expected size (567 bp for glmU and 504 bp for gyrB) were cut out and incubated with 50 µL of elution buffer (QIAamp DNA mini kit) overnight at 4°C. Extracted DNA was submitted for Sanger sequencing on an ABI 3730 DNA Analyzer (ThermoFisher Scientific) at Massey Genome Services (Palmerston North, NZ).
Molecular typing
Forward and reverse sequences were aligned to generate a consensus sequence in Geneious software, version 10.2.6 (Biomatters Inc., Auckland, NZ). Consensus sequences were aligned with reference sequences to assign species and serovars (Supplementary Table 3). Reference sequences included the glmU and gyrB regions of eight strains associated with two species, i.e. L. interrogans serovar Pomona str68, herein referred to as Pomona; L. interrogans serovar Copenhageni strain M20, herein referred to as Copenhageni; L. borgpetersenii serovar Hardjo type bovis strain I89, herein referred to as Hardjo type bovis; L. borgpetersenii serovar Balcanica strain Bal_possum, herein referred to as Balcanica_NZ; L. borgpetersenii serovar Balcanica strain Bal_Burgas, herein referred to as Balcanica_Burgas; L. borgpetersenii serovar Ballum strain Bal_mus127, herein referred to as Ballum; L. borgpetersenii serovar Tarassovi strain Perepelitsin, herein referred to as Tarassovi; and L. borgpetersenii strain Pacifica, herein referred to as Pacifica.
Ethics
This study was approved by the Royal (Dick) School of Veterinary Studies’ Veterinary Ethical Review Committee (reference number 15.19). According to Massey University's animal ethics committee guidelines, no additional ethical approval was required beyond that obtained for the original sampling as part of the previous projects.
Results
Molecular typing of Leptospira
A total of 997 samples were screened with the lipL32 qPCR, of which 211 tested positive and were subjected to the speciating PCR (). An additional 86 lipL32-negative samples were also tested with the speciating PCR because they had either tested positive to pathogenic Leptospira PCR in previous studies (n = 68) or were samples from animals with high titres against serovar Tarassovi in previous studies (n = 18). In total, 297 samples were tested with the speciating PCR, 148 samples were successfully amplified with the glmU PCR and 54 with the gyrB PCR (). A total of 202 amplicons were sent for sequencing, however 28 samples were amplified by both the glmU and gyrB PCR, thus 174 different samples were typed. Of the 174 amplicons, 123 were successfully typed following sequencing; 100 samples with the glmU PCR and 41 samples with the gyrB PCR, with 18 samples being typed with both PCR. Typing identified six strains/serovars belonging to two species: L. borgpetersenii strain Pacifica and serovars Balcanica_NZ, Ballum and Hardjo type bovis, and L. interrogans serovars Copenhageni and Pomona (). All generated sequences were identical to the reference strains and no new types were detected in any of the positive samples using the speciating PCR. All Balcanica sequences were determined by the glmU locus and were identical to the reference strain Bal_NZ, which has only been identified in New Zealand. Bal_possum has a single nucleotide polymorphism within the glmU region compared to the Balcanica_Burgas strain that was isolated from a human in Bulgaria (Wilkinson et al. Citation2021).
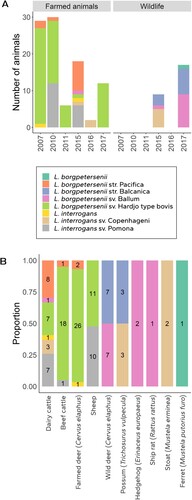
Figure 2. Leptospira species and serovars identified in DNA samples from New Zealand mammals stratified (A) by the year of sample acquisition and whether they were found in farmed animals or wildlife; and (B) by the mammalian hosts in which they were identified (numbers within each bar show the number of samples from each host in which each Leptospira strain/serovar was identified).
Host–Leptospira associations
Host–Leptospira associations were stratified by whether the host was farmed or wildlife over the study period (A). Farmed animals constituted dairy cattle, beef cattle, farmed deer, and sheep, while wildlife constituted wild red deer, brushtail possum, hedgehog, ship rat, stoat, mouse, and ferret. Farmed animals were positive for Hardjo type bovis (62/97; 64%), Pomona (18/97; 19%), Pacifica (11/97; 11%), Copenhageni (3/97; 3%) and Ballum (1/97; 1%). Wildlife was positive for Balcanica_NZ (10/26; 38%), Ballum (10/26; 38%) and Copenhageni (5/26; 19%).
Samples that were typed (n = 123), were also stratified by mammalian host and infecting Leptospira spp. (B). This stratification shows that each host type was positive to a range of different Leptospira spp. with dairy cattle being infected with five of the seven types detected in this study. One dairy cow had a mixed infection that limited typing to species level as L. interrogans, one deer sample could only be identified at species level as L. interrogans, and the ferret sample had poor-quality sequence data, limiting typing to species level as L. borgpetersenii (B).
Of the 11 samples that were positive for Pacifica in this study, six came from dairy cattle for which MAT data generated in previous studies were available. Four animals had titres to Tarassovi (1:96, 1:96, 1:768, and 1:1536); one animal had combined titres to Tarassovi (1:768), Copenhageni (1:48), Hardjo (1:48), and Ballum (1:24); and one animal had combined titres to Pomona (1:96), Copenhageni (1:96) and Hardjo (1:48). Samples without a positive titre were either not tested by MAT (one beef animal and two farmed deer) or only tested for Hardjo and Pomona (two dairy cattle).
Discussion
New Zealand has a high burden of leptospirosis in humans, which is atypical for a high-income nation with a temperate climate. A cornerstone for preventing disease in humans is the vaccination of pigs and dairy cattle (Marshall and Manktelow Citation2002). The combined cost of leptospirosis to the New Zealand economy including human disease (treatment and absence from work), loss of production in animals (cattle, sheep, and deer) and disease control measures (animal vaccinations) was estimated at $NZD24M annually (Sanhueza et al. Citation2020). Surveillance is an important tool for monitoring the Leptospira spp. in circulation and can have implications for disease control and vaccine development. In this study, we applied our recently developed molecular assay to historical samples, which allowed the correct identification of Leptospira species and serovars infecting farmed and wild mammals in New Zealand.
The molecular typing results revealed previously unidentified host–Leptospira associations in New Zealand, i.e. farmed deer infected with Pacifica (B). Farmed deer in New Zealand have largely been tested serologically for Hardjo type bovis, Pomona, Copenhageni, Ballum and Tarassovi (Subharat et al. Citation2012; Wilson et al. Citation2021). Although Tarassovi seropositivity is common in deer, molecular typing to identify the specific strain being shed was not available at the time the study was carried out (Wilson et al. Citation2021). As dairy cattle serologically positive to Tarassovi (Yupiana et al. Citation2019a) shed Pacifica (Wilkinson et al. Citation2021), previous serological classifications of Tarassovi in deer could likely be Pacifica. Pacifica has been considered an emerging public health risk for dairy farmers, as it was identified in dairy cattle samples collected in 2016 (Yupiana et al. Citation2019a) and environmental samples from a dairy farm in 2017 (Wilkinson et al. Citation2021). However, results from this study suggests that deer farmers could also be at risk from Pacifica. Thus, the Pacifica lineage may be more widespread than initially thought, as it has now been identified in two farming systems (B). Furthermore, Pacifica may not be an emerging strain, as it has been in circulation since at least 2007, as identified from the deer samples tested in this study (Supplementary Figure 1). It is difficult to ascertain how long Pacifica has been circulating in livestock as no samples from before 2007 were available for molecular typing. Since Pacifica has exclusively been found in farmed animals to date, it is likely maintained in livestock; however, only 7.1% (71/997) of the study's samples came from wildlife. A comprehensive survey of wildlife needs to be conducted to test this hypothesis.
We also found some new host–Leptospira associations in wildlife in New Zealand, including stoats and brushtail possums infected with Copenhageni, and a ferret infected with L. borgpetersenii (B). This finding is not surprising given the limited number of wild animals tested in New Zealand to date and the wide host range of leptospires in general. Globally, mustelids have been known to harbour Leptospira from serogroups Icterohaemorragiae (which includes serovar Copenhageni), as well as serogroups Australis, Autumnalis, Grippotyphosa and Sejroe (Ayral et al. Citation2016). Mustelids were first released in New Zealand in an attempt to control the rodent population (King Citation2017). Earlier studies show ship and brown rats harbouring Copenhageni in New Zealand (Brockie Citation1977; Carter and Cordes Citation1980) as well as other countries (Maas et al. Citation2018; Boey et al. Citation2019). The distribution of Leptospira infection in rats varies widely and has mainly been studied in urban environments (Boey et al. Citation2019). The infected brushtail possums and stoats in this study came from urban habitats that are shared with both ship and brown rats. Whether Copenhageni is maintained in both rats spp. and mustelids in New Zealand or whether there is inter-species transmission is difficult to say with the limited sample size in this study.
This study also reported a wide range of serovars associated with dairy cattle (B). These findings reinforce recent observations that serovar diversity in cattle is not just limited to serovars Hardjo type bovis and Pomona included in currently available vaccines (Yupiana et al. Citation2017, Citation2019b; Moinet Citation2020). While other livestock were infected with Hardjo type bovis, Pomona and Pacifica, dairy cattle were additionally infected with Ballum and Copenhageni, the two serovars that have been associated with wildlife in New Zealand (Moinet et al. Citation2021). In New Zealand, 99.5% of dairy farms vaccinate against serovars Hardjo type bovis and Pomona (Yupiana et al. Citation2017) and this has been shown to reduce shedding of the vaccinated strains (Yupiana et al. Citation2019b). However, this likely leaves an empty niche for new serovars. While historically dairy cattle were the recognised maintenance host for Hardjo type bovis and Pomona, these data indicate that Pacifica may also be maintained by dairy cattle. Human leptospirosis notification data (classified serologically) show an increase in Tarassovi cases in dairy farmers (Nisa et al. Citation2020). Due to lack of molecular typing, it is unclear if the genetic strain causing disease in dairy farmers is Tarassovi or Pacifica; however, molecular typing of dairy cattle in this study and the previous study (Yupiana et al. Citation2019a) only show evidence of Pacifica in dairy cattle with no genetic evidence of Tarassovi infection. The results from this study as well as the previous survey of dairy cattle (Yupiana et al. Citation2019a) suggest a relationship between Tarassovi seropositivity and Pacifica infection where both likely belong to serogroup Tarassovi. However, this can only be determined upon isolation and characterisation of strain Pacifica and all attempts to isolate this strain to date have been unsuccessful. Serological testing with Tarassovi before the availability of molecular typing tools explains why Pacifica remained undescribed prior to 2019 (Yupiana et al. Citation2019a).
In previous studies, mice, rats, and hedgehogs were found to harbour serovar Ballum, where mice were considered the primary maintenance host and rats and hedgehogs as secondary maintenance hosts (Brockie Citation1977; Moinet Citation2020; Moinet et al. Citation2021). In this study, hedgehogs and rats were infected with Ballum, but so were wild red deer (B). This is the first report of Ballum infection in wild red deer, but it is unclear if wild red deer are a maintenance host or spill-over host from being in the same environment with mice, rats and hedgehogs. Similarly, while brushtail possums have been known to be the maintenance host for Balcanica_NZ in New Zealand (Hathaway et al. Citation1978), this is the first report of Balcanica_NZ in wild red deer. A comprehensive survey needs to be conducted to determine if wild red deer can maintain Ballum and Balcanica_NZ, as sample numbers in this study were low. Overall, the wild mammals in New Zealand appear to maintain Balcanica_NZ, Ballum and Copenhageni while farmed animals appear to be the maintenance host for Hardjo type bovis, Pomona and Pacifica, except for dairy cattle, which are also infected with wildlife-associated serovars Ballum and Copenhageni. The results from this study also verified that the Hardjo type bovis in farmed animals that were previously typed serologically, and are serologically indistinct from Balcanica, were indeed Hardjo type bovis and not Balcanica. Lastly, the results of this study also reconfirmed previously described host–Leptospira associations including dairy cattle infected with Copenhageni, Pomona, Ballum, Hardjo type bovis and strain Pacifica; sheep infected with Pomona and Hardjo type bovis; brushtail possum infected with Balcanica_NZ; farmed deer infected with Hardjo type bovis; and hedgehogs infected with Ballum (B).
While this study revealed some new host–Leptospira associations, there are several limitations. Firstly, not all samples from the previous studies were tested in this study, i.e. samples that were previously determined to be negative for pathogenic Leptospira using PCR were excluded from this study unless they met selection criteria for typing 3 (). This was to conserve resources on samples known to be negative. Thus, this convenience sampling and a low number of available samples for some species prevented an accurate estimation of prevalence of the different serovars in their respective mammalian species. Further research with appropriate sampling methods, e.g. investigating the difference in serovars from vaccinated and unvaccinated dairy cattle, or wildlife from rural and urban regions, within a defined period and with larger sample sizes is necessary to estimate prevalence. Secondly, typing in this study was often limited by the bacterial load in the samples as determined by the Cq values of the qPCR, where a higher Cq value denoted lower bacterial load. Of the 211 samples positive with the lipL32 qPCR, 103 were typed, of which 32% (33/103) had Cq values of ≥ 30 while of the 108 samples that were not typed, 90% (97/108) had Cq values of ≥ 30. Thus, while samples with lower bacterial load were positive, the quantity of DNA was insufficient to provide quality sequence for typing, therefore decreasing the number of positive samples that could be typed.
In conclusion, this study has updated understanding of host–Leptospira associations in New Zealand (). Many studies in the last decade have focused solely on Hardjo type bovis and Pomona with reliance on serology (Dorjee et al. Citation2008; Vallée et al. Citation2015; Dreyfus et al. Citation2018) and very few studies have investigated Ballum, Copenhageni and Tarassovi (Yupiana et al. Citation2019a; Moinet Citation2020; Wilson et al. Citation2021). While serology remains a useful tool, it is not uncommon for seronegative animals to shed leptospires (Brockie Citation1977; Hathaway et al. Citation1981) and there can be cross-reactivity and inaccurate classification. Future surveillance and research will need to incorporate molecular typing and will need to focus on a wider diversity of serovars to better understand their distributions, prevalence, and overall impacts.
Table 2. An updated summary of Leptospira spp. detected by culture or molecular typing in New Zealand mammals.
Supplemental Material
Download PDF (601.1 KB)Acknowledgements
We would like to thank Neville Haack, Emilie Vallée, Yuni Yupiana, Jennifer Parramore, Ruth Meenks, Supatsak Subharat, Mark van de Pol, Fang Fang, Grace Miller, Bernard Bangham, Daniel Ritchie, and David Wiessing for collecting samples from the mammals tested in this study, and Julie Collins-Emerson and Cord Heuer for supervision of the students. This project was funded by the Palmerston North Medical Research Foundation [grant number 20817] and supported by a Massey University SOVS Summer Scholarship.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ayral F, Djelouadji Z, Raton V, Zilber AL, Gasqui P, Faure E, Baurier F, Vourc’h G, Kodjo A, Combes B. Hedgehogs and mustelid species: major carriers of pathogenic Leptospira, a survey in 28 animal species in France (2012–2015). PLoS ONE 11, e0162549, 2016. https://doi.org/10.1371/journal.pone.0162549
- Babudieri B. Proposed standardization of the agglutination-adsorption test for Leptospira. Bulletin of the World Health Organization 44, 795–810, 1971
- Bahaman AR, Marshall RB, Blackmore DK, Hathaway SW. Isolation of Leptospira interrogans serovar Hardjo from sheep in New Zealand. New Zealand Veterinary Journal 28, 171, 1980. https://doi.org/10.1080/00480169.1980.34738
- Benschop J, Nisa S, Spencer SEF. Still ‘dairy farm fever’? A Bayesian model for leptospirosis notification data in New Zealand. Journal of the Royal Society Interface 18, 20200964, 2021. https://doi.org/10.1098/rsif.2020.0964
- *Bingham P. Quarterly report of investigations of suspected exotic diseases: July to September 2021. Surveillance 48(4), 10–24, 2021
- Boey K, Shiokawa K, Rajeev S. Leptospira infection in rats: a literature review of global prevalence and distribution. PLoS Neglected Tropical Diseases 13, e0007499, 2019. https://doi.org/10.1371/journal.pntd.0007499
- *Bolt I. Leptospirosis in New Zealand pig herds: an epidemiological study and a computer simulation model of endemic leptospiral infection in New Zealand with particular reference to Leptospira interrogans serovar Pomona. PhD thesis, Massey University, Palmerston North, NZ, 1990
- Brockie RE. Leptospiral infections of rodents in the North Island. New Zealand Veterinary Journal 25, 89–96, 1977. https://doi.org/10.1080/00480169.1977.34369
- Carter ME, Cordes DO. Leptospirosis and other infections of Rattus rattus and Rattus norvegicus. New Zealand Veterinary Journal 28, 45–50, 1980. https://doi.org/10.1080/00480169.1980.34688
- Chideroli RT, Gonçalves DD, Suphoronski SA, Alfieri AF, Alfieri AA, de Oliveira AG, de Freitas JC, Pereira UP. Culture strategies for isolation of fastidious Leptospira serovar Hardjo and molecular differentiation of genotypes Hardjobovis and Hardjoprajitno. Frontiers in Microbiology 8, 2155, 2017. https://doi.org/10.3389/fmicb.2017.02155
- Dorjee S, Heuer C, Jackson R, West DM, Collins-Emerson JM, Midwinter AC, Ridler AL. Prevalence of pathogenic Leptospira spp. in sheep in a sheep-only abattoir in New Zealand. New Zealand Veterinary Journal 56, 164–70, 2008. https://doi.org/10.1080/00480169.2008.36829
- Dreyfus A, Wilson P, Benschop J, Collins-Emerson J, Verdugo C, Heuer C. Seroprevalence and herd-level risk factors for seroprevalence of Leptospira spp. in sheep, beef cattle and deer in New Zealand. New Zealand Veterinary Journal 66, 302–11, 2018. https://doi.org/10.1080/00480169.2018.1507770
- Dunay SN, Bass JS, Stremick J. Leptospirosis: a global health burden in review. Emergency Medicine 6, 336, 2016. https://doi.org/10.4172/2165-7548.1000336
- *Fairly R. Porcine leptospirosis in New Zealand. Surveillance 24(4), 15, 1997
- Fang F, Collins-Emerson JM, Heuer C, Hill FI, Tisdall DJ, Wilson PR, Benschop J. Interlaboratory and between-specimen comparisons of diagnostic tests for leptospirosis in sheep and cattle. Journal of Veterinary Diagnostic Investigation 26, 734–47, 2014. https://doi.org/10.1177/1040638714548476
- Hathaway SC, Blackmore DK, Marshall RB. The serologic and cultural prevalence of Leptospira interrogans serovar Balcanica in possums (Trichosurus vulpecula) in New Zealand. Journal of Wildlife Diseases 14, 345–50, 1978. https://doi.org/10.7589/0090-3558-14.3.345
- Hathaway SC, Blackmore DK, Marshall RB. Leptospirosis in free-living species in New Zealand. Journal of Wildlife Diseases 17, 489–96, 1981. https://doi.org/10.7589/0090-3558-17.4.489
- King CM. Liberation and spread of stoats (Mustela erminea) and weasels (M. nivalis) in New Zealand, 1883–1920. New Zealand Journal of Ecology 41, 163–77, 2017. https://doi.org/10.20417/nzjecol.41.29
- Korba AA, Lounici H, Kainiu M, Vincent AT, Mariet JF, Veyrier FJ, Goarant C, Picardeau M. Leptospira ainlahdjerensis sp. nov., Leptospira ainazelensis sp. nov., Leptospira abararensis sp. nov. and Leptospira chreensis sp. nov., four new species isolated from water sources in Algeria. International Journal of Systematic and Evolutionary Microbiology 71, 2021. https://doi.org/10.1099/ijsem.0.005148
- Maas M, De Vries A, Reusken C, Buijs J, Goris M, Hartskeerl R, Ahmed A, Van Tulden P, Swart A, Pijnacker R, et al. Prevalence of Leptospira spp. and Seoul hantavirus in brown rats (Rattus norvegicus) in four regions in the Netherlands, 2011–2015. Infection Ecology & Epidemiology 8, 1490135, 2018. https://doi.org/10.1080/20008686.2018.1490135
- Mackintosh CG, Blackmore DK, Marshall RB. Isolation of Leptospira interrogans serovars Tarassovi and Pomona from dogs. New Zealand Veterinary Journal 28, 100, 1980a. https://doi.org/10.1080/00480169.1980.34709
- Mackintosh CG, Marshall RB, Blackmore DK. Leptospira interrogans serovar Balcanica in cattle. New Zealand Veterinary Journal 28, 268, 1980b. https://doi.org/10.1080/00480169.1980.34774
- *Marshall R, Chereshsky A. Vaccination of dairy cattle against leptospirosis as a means of preventing human infections. Surveillance 23(1), 27–8, 1996
- Marshall R, Manktelow B. Fifty years of leptospirosis research in New Zealand: a perspective. New Zealand Veterinary Journal 50, 61–3, 2002. https://doi.org/10.1080/00480169.2002.36270
- *Moinet M. Molecular and eco-epidemiology of Leptospira borgpetersenii serovar Ballum in wild invasive mammals in a farming environment in New Zealand. PhD thesis, Massey University, Palmerston North, NZ, 2020
- Moinet M, Wilkinson DA, Aberdein D, Russell JC, Vallée E, Collins-Emerson JM, Heuer C, Benschop J. Of mice, cattle, and men: a review of the eco-epidemiology of Leptospira borgpetersenii serovar Ballum. Tropical Medicine and Infectious Disease 6, 189, 2021. https://doi.org/10.3390/tropicalmed6040189
- Musso D, La Scola B. Laboratory diagnosis of leptospirosis: a challenge. Journal of Microbiology, Immunology and Infection 46, 245–52, 2013. https://doi.org/10.1016/j.jmii.2013.03.001
- Nisa S, Wilkinson DA, Angelin-Bonnet O, Paine S, Cullen K, Wright J, Baker MG, Benschop J. Diverse epidemiology of Leptospira serovars notified in New Zealand, 1999–2017. Pathogens 9, 841, 2020. https://doi.org/10.3390/pathogens9100841
- Picardeau M. Virulence of the zoonotic agent of leptospirosis: still terra incognita? Nature Reviews Microbiology 15, 297–307, 2017. https://doi.org/10.1038/nrmicro.2017.5
- Ramadass P, Marshall RB, Jarvis BD. Species differentiation of Leptospira interrogans serovar Hardjo strain Hardjobovis from strain Hardjoprajitno by DNA slot blot hybridisation. Research in Veterinary Science 49, 194–7, 1990. https://doi.org/10.1016/S0034-5288(18)31076-2
- Ris DR, Lake DE, Holland JTS. The isolation of Leptospira serotypes Copenhageni and Ballum from healthy calves. New Zealand Veterinary Journal 21, 218–20, 1973. https://doi.org/10.1080/00480169.1973.34111
- Robinson AJ, Ramadass P, Lee A, Marshall RB. Differentiation of subtypes within Leptospira interrogans serovars Hardjo, Balcanica and Tarassovi, by bacterial restriction-endonuclease DNA analysis (BRENDA). Journal of Medical Microbiology 15, 331–8, 1982. https://doi.org/10.1099/00222615-15-3-331
- Ryan TJ, Marshall RB. Isolation of a leptospire belonging to serogroup Tarassovi. New Zealand Veterinary Journal 24, 212–3, 1976. https://doi.org/10.1080/00480169.1976.34320
- Sanhueza JM, Baker MG, Benschop J, Collins-Emerson JM, Wilson PR, Heuer C. Estimation of the burden of leptospirosis in New Zealand. Zoonoses and Public Health 67, 167–76, 2020. https://doi.org/10.1111/zph.12668
- Schollum LM, Blackmore DK. The serological and cultural prevalence of leptospirosis in a sample of feral goats. New Zealand Veterinary Journal 29, 104–6, 1981. https://doi.org/10.1080/00480169.1981.34813
- Slack AT, Symonds ML, Dohnt MF, Smythe LD. Identification of pathogenic Leptospira species by conventional or real-time PCR and sequencing of the DNA gyrase subunit B encoding gene. BMC Microbiology 6, 95, 2006. https://doi.org/10.1186/1471-2180-6-95
- Stoddard RA, Gee JE, Wilkins PP, McCaustland K, Hoffmaster AR. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagnostic Microbiology and Infectious Disease 64, 247–55, 2009. https://doi.org/10.1016/j.diagmicrobio.2009.03.014
- *Subharat S. Epidemiology, diagnosis and vaccination control of leptospirosis in farmed deer in New Zealand. PhD thesis, Massey University, Palmerston North, NZ, 2010
- Subharat S, Wilson P, Heuer C, Collins-Emerson J. Longitudinal serological survey and herd-level risk factors for Leptospira spp. serovars Hardjo-bovis and Pomona on deer farms with sheep and/or beef cattle. New Zealand Veterinary Journal 60, 215–22, 2012. https://doi.org/10.1080/00480169.2012.663323
- Vallée E, Heuer C, Collins-Emerson JM, Benschop J, Wilson PR. Serological patterns, antibody half-life and shedding in urine of Leptospira spp. in naturally exposed sheep. New Zealand Veterinary Journal 63, 301–12, 2015. https://doi.org/10.1080/00480169.2015.1049668
- Vincent AT, Schiettekatte O, Goarant C, Neela VK, Bernet E, Thibeaux R, Ismail N, Mohd Khalid MKN, Amran F, Masuzawa T, et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Neglected Tropical Diseases 13, e0007270, 2019. https://doi.org/10.1371/journal.pntd.0007270
- Wilkinson DA, Edwards M, Benschop J, Nisa S. Identification of pathogenic Leptospira species and serovars in New Zealand using metabarcoding. PLoS ONE 16, e0257971, 2021. https://doi.org/10.1371/journal.pone.0257971
- Wilson PR, Mannewald A, Collins-Emerson JM, Dreyfus A, Sanhueza JM, Benschop J, Verdugo C, Emanuelson U, Boqvist S, Heuer C. Serological study of Leptospira interrogans serovar Copenhageni and L. borgpetersenii serovars Tarassovi and Ballum in beef cattle, sheep and deer in New Zealand. New Zealand Veterinary Journal 69, 83–92, 2021. https://doi.org/10.1080/00480169.2020.1830867
- *Yupiana Y, Collins-Emerson J, Benschop J, Weston J, Wilson P, Vallée E, Heuer C. Nationwide survey of leptospiral antibodies and shedding in New Zealand dairy herds. In: Conference Proceedings of the Society of Dairy Cattle Veterinarians of the NZVA. Pp 24–5. New Zealand Veterinary Association, Wellington, NZ, 2017
- Yupiana Y, Vallée E, Wilson P, Collins-Emerson J, Weston J, Benschop J, Heuer C. Emerging Leptospira strain poses public health risk for dairy farmers in New Zealand. Preventive Veterinary Medicine 170, 104727, 2019a. https://doi.org/10.1016/j.prevetmed.2019.104727
- Yupiana Y, Wilson PR, Weston JF, Vallée E, Collins-Emerson JM, Benschop J, Scotland T, Heuer C. Epidemiological investigation of Leptospira spp. in a dairy farming enterprise after the occurrence of three human leptospirosis cases. Zoonoses and Public Health 66, 470–9, 2019b. https://doi.org/10.1111/zph.12578
- *Non-peer-reviewed
