Abstract
Waxflower is one of Australia's major native cut flowers for the export market. A number of interspecific hybrid cultivars such as the ‘Pearl’ series bred by the Department of Agriculture and Food, Western Australia have increased the competitiveness of waxflower on world markets. To improve the breeding efficiency, resistance gene analog polymorphisms (RGAP) are investigated as molecular markers for the early identification of interspecific hybrids between Chamelaucium uncinatum and C. megalopetalum. The results show that RGAP can be effectively applied to generate DNA markers to identify true waxflower hybrids. The RGAP marker system provides a reliable, simple, fast and inexpensive approach for hybrid identification in waxflower breeding.
Introduction
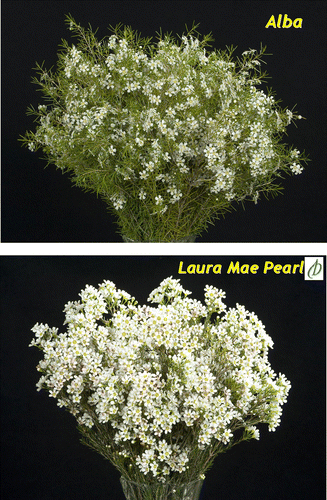
Waxflower (Chamelaucium Desf.; Myrtaceae) is one of the key Australian native flowers for export (Shan & Seaton Citation2009). The development of the Australian waxflower industry relies on the continual supply of new varieties (Shan & Seaton Citation2008). To meet this requirement, an extensive waxflower breeding programme is under way at the Department of Agriculture and Food, Western Australia (DAFWA). This breeding programme has released eight interspecific hybrids and two intergeneric hybrids so far. These hybrids have improved cut flower characteristics in terms of form (), colour, vase life and flower bud shattering (Seaton & Poulish Citation2010).
Fig. 1 Comparison of Chamelaucium uncinatum variety Alba with interspecific hybrid Laura Mae Pearl![]()
Although emasculation has been practised, the likelihood of producing false hybrids from selfing or pollen cross contamination exists. However, a true hybrid's identity is critical for its plant breeder's right (PBR) and patent application if released as a new variety. In addition, false hybrids present a costly problem taking up valuable resource and effort in propagation, planting out and maintenance until they flower when evaluation can be conducted. Therefore development of a simple, reliable, inexpensive and fast method to identify true hybrids at an early stage, such as at in vitro stage or as young seedlings, would allow false hybrids to be discarded before planting out, leading to higher breeding efficiency.
Morphological markers can be used to identify true hybrids (Oliveira et al. 2002). However, it is not easy in most cases to find a distinguishable marker, particularly at early seedling stage. In addition, morphological characters are subject to environmental influence, which adds to the difficulty of identification.
Molecular markers are good alternatives for identifying hybrids in plants as these markers are DNA based, and are not subject to environmental influences. Many molecular markers have been applied for parentage discrimination in hybrids and identification of hybrids including random amplified polymorphism DNA (RAPD), simple sequence repeat (SSR), PCR-based restriction fragment length polymorphism (PCR-RFLP), amplified fragment length polymorphism (AFLP) and random amplified microsatellite polymorphism (RAMP) (Dubouzet et al. Citation1998; Oliveira et al. Citation2002; Kiew et al. Citation2003; Bateman & Hollingsworth Citation2004; Khasa et al. Citation2005; Lee & Han Citation2006; Liu et al. Citation2007; Kim et al. Citation2008). Among these markers, RAPD has the advantage of low cost, being fast and simple, but the application is limited due to its poor reproducibility when amplification conditions change. SSR provides reliable co-dominant markers, but the cost on development of microsatellite loci in a new crop is very high. AFLP enjoys reputation of abundance in polymorphism and high reproducibility, but technical complexity and requirement of expensive equipment limits its application in some circumstances. PCR-RFLP is relatively inexpensive and fast (Harry et al. Citation1998) due to its PCR nature, but lack of sufficient diversity is its drawback (Liu et al. Citation2007). RAMP can detect large amounts of polymorphism in a single reaction and is highly reproducible, but the cost is higher and technically difficult compared with PCR-RFLP (Liu et al. Citation2007).
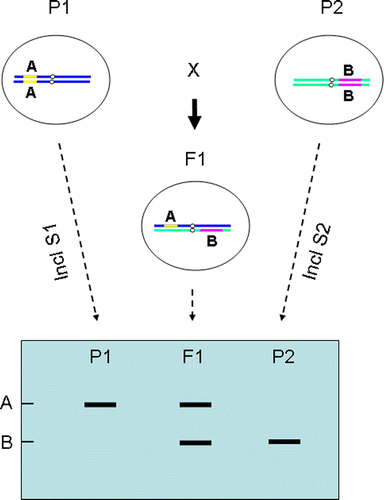
The cloning and sequencing of disease resistance genes (R genes) from diverse plant species have found that many R genes contain a well-conserved DNA sequence called nucleotide binding sites (NBS) (Yu et al. Citation1996; Young Citation2000; Lacock et al. Citation2003). In recent years, NBS sequences have been used to design oligo primers for PCR to amplify and detect resistance gene analog polymorphisms (RGAP) (Shi et al. Citation2001; Wen et al. Citation2008). Most NBS-derived primers contain 18 to 24 nucleotides (Collins et al. Citation1998; Lopez et al. Citation2003), which enable RGAP to be generated in much more stringent conditions in the PCR than other DNA fingerprinting methods where short oligo primers are employed, such as RAPD (Williams et al. Citation1990). Since NBS sequences are abundant in plant genomes (Meyers et al. Citation1999), NBS-derived primers amplify multiple DNA fragments in each PCR (Shi et al. Citation2001; Wen et al. Citation2008). Although NBS-primer amplified DNA polymorphisms may, or may not, be related to plant disease resistance genes (Zhu et al. Citation2002), RGAP provides a fast, easy and reliable DNA fingerprinting approach, which can be used to compare the genetic diversity at the DNA level between individual plants (Diaz & Ferrer Citation2003). The RGAP may also be applicable in the DAFWA waxflower breeding programme for the identification of interspecific waxflower hybrids according to the following hypothesis (). When the diploid parent plants are genetically homozygous, the polymorphic DNA bands from parents will be inherited and presented in all the hybrids, which is true if either the DNA bands are dominant, i.e., from different loci in the genome (as shown in ), or if the DNA bands are co-dominant and the allelic bands are from the same locus.
The aim of this research was to determine whether the DNA marker system RGAP could be applied for routine identification and selection of true hybrids at the early growing stage in the waxflower breeding programme.
Materials and methods
Crosses and interspecific hybrids
Crosses and reciprocal crosses were made between Chamelaucium uncinatum and C. megalopetalum. The breeding line of C. uncinatum used in this study has bright purple flowers with a white central ring. The breeding line of C. megalopetalum used in this study has white flowers. The progeny were derived from strictly controlled crosses with emasculation to avoid selfing and cross contaminations. Progeny embryos were rescued 4–5 weeks after artificial pollination and seedlings were clonally propagated using plant tissue culture procedure established at DAFWA (Shan & Seaton Citation2009). A total of 26 plants were grown to maturity at Medina Research Station and DAFWA campus at South Perth and selected for analysis.
Plants tested were:
* C. uncinatum parent (P1), five plants (numbered 1–5)
* C. megalopetalum parent (P2), three plants (numbered 6–8)
* the cross C. uncinatum×C. megalopetalum (P1×P2) produced four progeny: H1 (plants numbered 9–13); H2 (plants numbered 14–15); H3 (plant numbered 16); and H4 (plant numbered 17)
* the reciprocal cross C. megalopetalum×C. uncinatum (P2×P1) produced three progeny: RH1 (plants numbered 18–20); RH2 (plant numbered 21); and RH3 (plant numbered 22)
* P1 self, two plants (numbered 23–24)
* P2 self, two plants (numbered 25–26).
Generation and detection of RGAP markers
DNA was extracted from young leaves of the above plants using a Nucleon Phytopure Extraction Kit based on the protocol provided (Amersham Biosciences). Two pairs of NBS-sequence derived primers () were adopted from You et al. (Citation2004). RGAP was conducted in 10 µl PCR mix containing about 50 pg of plant genomic DNA, 1 unit of Taq polymerase (Fisher Biotec, Perth), 6 pmol each of either the primer pair P loop-F and GLPL1, or the primer pair P loop-F and Kin2-A, 6 pmol MseI-primer, 67 mM Tris-HCl (pH8.8), 2 mM MgCl2, 16.6 mM (NH4)2SO4, 0.45% Triton X-100, 4 µg gelatin and 0.2 mM dNTPs. The primer P loop-F was labelled with γ-33P as described by Yang et al. (Citation2001) . PCR was cycled on a Hybaid DNA Express thermocycler for 40 cycles each of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min. The amplified products were resolved on a manual sequencing gel using a Sequi-Gen GT sequencing cell (Bio-Rad) at 55 W for 3 h. RGAP image was detected by autoradiography using an X-ray film (Yang et al. Citation2001).
Table 1 NBS-sequence derived primer sequences
Morphological examination and confirmation of true hybrids to validate the established RGAP marker system
When the plants reached flowering stage, morphological characteristics of the progeny and their parents were examined. Flowering time was recorded. The colours of flowers, buds and flower receptacles were determined from colour charts produced by the Royal Horticultural Society (5th edition). Leaf shape was described. Leaf length and flower size was measured. Analysis of variance (ANOVA) on leaf length and flower size was carried out using GenStat 12.1 (PC/Windows XP). A LSD (least significant difference) value at 5% level was used to compare all the progenies with both P1 and P2. All these characteristics were used as criteria to confirm hybrid identity.
Results and discussion
Detection of RGAP markers to identify hybrids
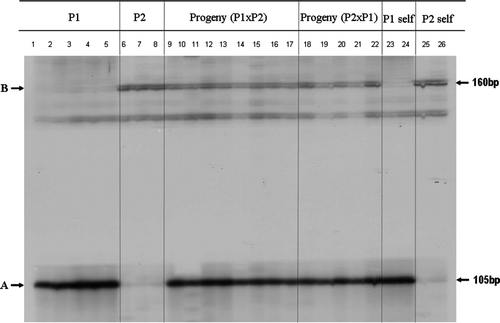
The markers specific to the genotypes of P1 and P2 were found from the RGAP PCR products generated by the primer pair of P loop-F and GLPL1 (). The marker of 105 bp is specific to P1 (lane 1 to 5). The marker of 160 bp is specific to P2 (lane 6 to 8). Both markers were present in the progeny either with P1 as mother (lane 9 to 17) or with P2 as mother (lane 18 to 22). These RGAP markers behaved in the same manner assumed in the hypothesis (, ) although we were not sure if these markers came from the same locus or different loci. From the band intensity it indicated that the two RGAP markers might have originated from different loci; however, it was also possible that the two markers might be from different alleles of the same locus. It was interesting, but not surprising, that the two RGAP markers (arrowed in ) showed a different intensity even if they were two allelic DNA bands from the same locus. In our earlier work with lupin (Lupinus angustifolius L), we found that one of three allelic MFLP markers linked to a disease resistance gene was weaker than the other two allelic bands from the same locus (Yang et al. Citation2008), where DNA sequencing found that one of the DNA primers used PCR did not have a perfect match with the template DNA, making DNA amplification less efficient which resulted in a weaker DNA band (Yang et al. Citation2008).
Fig. 3 Gel image of RGAP DNA markers A and B generated by P loop-F and GLPL1 primers (P1: parent C. uncinatum; P2: parent C. megalopetalum).
However, as with other markers like RAMP (Liu et al. Citation2007), not all NBS-sequence derived primers could detect the male- and female-specific fragments simultaneously. The primer combination of P loop-F and Kin2-A did not reveal markers specific to P1 although it amplified a band of 240 bp specific to P2. This band was present in both P2 and F1 progeny. This indicated that some other primer combinations might not detect any specific markers to both parents. Therefore, it is necessary that the parental lines in the crosses should be screened with various primer combinations before a suitable primer pair is selected for implementation on hybrid identification.
Morphological validation of the RGAP marker systems
All the progeny used in this study were morphologically determined as true hybrids and the best characteristics for confirmation of the hybrids were petal and receptacle colours, leaf shape, leaf and flower size (). Compared with P1, all progeny (H1 to H4 and RH 1 to RH3) had white petal colour with or without a different shade of purple, which was different from the purple in P1. All progeny had flower receptacles in yellow-green but purple in P1. Compared with P2, all progeny had narrow linear leaves different from elliptical in P2 and leaves in the progeny were 33% to 43% longer than the P2 (P < 0.05). In addition, all the flowers in the progeny were 4.5% to 21.2% bigger than P2 in diameter (P < 0.05).
Table 2 Biodiversity of waxflower progeny and their parents
Other characteristics were not good enough to act as morphological markers to identify hybrids, such as flowering time, flower bud and leaf colour, although there were variations between the samples examined. All the progeny had a similar flowering period of time as their parents, although some started earlier, as in H3, or a little later than P1, like RH3. All the progeny had flower buds in grey-orange and leaves in green group.
RGAP markers can be used to identify the true hybrids in plant breeding programme
The validation of the true hybrids by morphological characters indicated that RGAP markers, such as markers A and B specific to P1 and P2 respectively for Chamelaucium plants in this study (), could be used for hybrid identification.
The method using RGAP markers to identify hybrids was found to be reliable. It is necessary when using this method to have at least two specific markers with the same primer combination. One marker is specific to the female parent line and the other to the pollen donor parental line. In this case, the presence of both specific markers served as a proof of the true hybrid (). Only one marker specific to either single parent is not enough, for example, the 240 bp marker generated by primers P loop-F and Kin2-A.
Using RGAP markers in the method presented here was accurate in the identification of true hybrids in waxflowers where little or no genome information was available. It also lends itself to routine use in a breeding programme for waxflowers.
The identification can be done at a very early stage of plant growth. Only 50–100 mg of young leaves is required for the DNA analysis, which is quite easy to achieve from in vitro micro-cuttings. The examination can be done within a couple of days from the DNA extraction to the identification of hybrid identity. Dozens of samples can be assessed easily per person per day.
This method is inexpensive and feasible. Since the method is based on PCR reaction, basic equipment can meet the evaluation requirements. This method is available to breeders who are not able to access expensive equipment, such as a DNA sequencer which is required for the generation of AFLP markers. Breeders who have no access to radioisotope facilities can use fluorescent dye to label the primer in PCR; our earlier work with lupins has shown that replacing 33P with florescence labelling would achieve the same successful results (Yang et al. Citation2004; You et al. Citation2004).
RGAP can be employed not only to detect resistant genes (Shi et al. Citation2001; Wen et al. Citation2008), but also can be applied to generate DNA markers to identify true hybrids. The RGAP marker system established here provides a practical approach for hybrid identification in a plant breeding programme.
Acknowledgements
We thank Chris McMullan and George Morris for crossing and growing plants; Jan Hopper for early embryo rescue of the hybrids; Mario D'Antuono for his help with data analysis, and Medina Research Station staff for maintaining our plants. We gratefully thank the referees for their valuable suggestions.
References
- Bateman , RM and Hollingsworth , PM . 2004 . Morphological and molecular investigation of the parentage and maternity of Anacamptis×albuferensis (A. fragrans×A. robust), a new hybrid orchid from Mallorca . Spain Taxon , 53 : 43 – 54 .
- Collins , NC , Webb , CA , Seah , S , Ellis , JG , Hulbert , SH and Pryor , A . 1998 . The isolation and mapping of disease resistance gene analogs in maize . Molecular Plant-Microbe Interactions , 10 : 968 – 978 .
- Diaz , V and Ferrer , E . 2003 . Genetic variation of population of Pinus cocarpa revealed by resistance analog polymorphisms . Genome , 46 : 404 – 410 .
- Dubouzet , JG , Shinoda , K and Murata , N . 1998 . Interspecific hybridization of Allium giganteum Regel: production and early verification of putative hybrids . Theoretical and Applied Genetics , 96 : 385 – 388 .
- Harry , DE , Temesgen , B and Neale , DB . 1998 . Codominant PCR-based markers for Pinus taeda developed from mapped cDNA clones . Theoretical and Applied Genetics , 97 : 327 – 336 .
- Khasa , D , Pollefeys , P , Navarro-Quezada , A , Perinet , P and Bousquet , J . 2005 . Species-specific microsatellite markers to monitor gene flow between exotic populars and their natural relatives in eastern North America . Molecular Ecology Notes , 5 : 920 – 923 .
- Kiew , R , Teo , LL and Gan , YY . 2003 . Assessment of the hybrid status of some malesian plants using amplified fragment length polymorphism . Telopea , 10 : 225 – 233 .
- Kim , G-J , Gi , G-Y , Lee , J-H , Joung , Y-H , Song , Y-H and Han , T-H . 2008 . Application of RAPD marker for the detection of parentage from known rose pedigrees . Horticulture, Environment, and Biotechnology , 49 : 253 – 257 .
- Lacock , L , Niekerk , C , Loots , S , Preez , F and Botha , AM . 2003 . Functional and comparative analysis of expressed sequences from Diuraphis noxia infested wheat obtained utilising the conserved nucleotide binding site . African Journal of Biotechnology , 2 : 75 – 81 .
- Lee , JH and Han , TH . 2006 . Identification of parental species of the Alstroemeria cv. ‘Jubilee’ using AFLP marker technique . Scientia Horticulturae , 111 : 63 – 67 .
- Liu , H , Yan , G , Finnegan , PM and Sedgley , R . 2007 . Development of DNA markers for hybrid identification in Leucadendron (proteaceae) . Scientia Horticulturae , 113 : 376 – 382 .
- Lopez , CE , Acosta , IF , Jara , C , Pedraza , F , Baitan-Solis , E , Gallego , G , Beebe , S and Tohme , J . 2003 . Identifying resistance gene analogs associated to different pathogens in common bean . Phytopathology , 93 : 88 – 95 .
- Meyers , BC , Dickerman , AW , Michelmore , RW , Sivaramakrishnan , S , Sobral , BW and Young , ND . 1999 . Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily . Plant Journal , 20 : 317 – 332 .
- de Oliveira , A , Novac , GA , Cristofani , M and Machado , M . 2002 . Identification of citrus hybrids through the combination of leaf apex morphology and SSR markers . Euphytica , 128 : 397 – 403 .
- Seaton K , Poulish N 2010 . Production of premium waxflowers . Department of Agriculture and Food, Western Australia. Bulletin 4778 .
- Shan F , Seaton K 2008 . Remaining competitive through innovative breeding of Australian native flowers . Horticulture Fifth Workshop, The Department of Agriculture and food western Australia . 42 43 .
- Shan , F and Seaton , K . 2009 . Enhancing waxflower breeding efficiency through early embryo rescue . Acta Hortculturae , 829 : 183 – 187 .
- Shi , ZX , Chen , XM , Line , RF , Leung , H and Wellings , CR . 2001 . Development of resistance gene analog polymorphism markers for the Yr9 gene resistance to wheat stripe rust . Genome , 44 : 509 – 516 .
- Wen , W , Li , G , He , Z , Yang , W , Xu , M and Xia , X . 2008 . Development of an STS marker tightly linked to Yr26 against wheat stripe rust using the resistance gene-analog polymorphism (RGAP) technique . Molecular Breeding , 22 : 507 – 515 .
- Williams , JGK , Kubelik , ARK , Livak , JL , Rafalski , JA and Tingey , SV . 1990 . DNA polymorphisms amplified by random primers are useful as genetic markers . Nucleic Acids Research , 18 : 6531 – 6535 .
- Yang , H , Boersma , JG , You , M , Buirchell , BJ and Sweetingham , MW . 2004 . Development and implementation of a sequence-specific PCR marker linked to a gene conferring resistance to anthracnose disease in narrow-leafed lupin (Lupinus angustifolius L.) . Molecular Breeding , 14 : 145 – 151 .
- Yang , H , Renshaw , D , Thomas , G , Buirchell , BJ and Sweetingham , MW . 2008 . A strategy to develop molecular markers applicable to a wide range of crosses for marker assisted selection in plant breeding: a case study on anthracnose disease resistance in lupin (Lupinus angustifolius L.) . Molecular Breeding , 21 : 473 – 483 .
- Yang , H , Sweetingham , MW , Cowling , WA and Smith , PMC . 2001 . DNA fingerprinting based on microsatellite-anchored fragment length polymorphisms, and isolation of sequence-specific PCR markers in lupin (Lupinus angustifolius L.) . Molecular Breeding , 7 : 203 – 209 .
- You , M , Boersma , JG , Buirchell , BJ , Sweetingham , MW , Siddique , KHM and Yang , H . 2004 . A PCR-based molecular marker applicable for marker-assisted selection for anthracnose disease resistance in lupin breeding . Cellular and Molecular Biology Letters , 10 : 123 – 134 .
- Young , ND . 2000 . The genetic architecture of resistance . Current Opinion in Plant Biology , 3 : 285 – 290 .
- Yu YG Buss GR Saghai Maroof MA Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binging site Proceedings of the National Academy of Sciences 1996 93 11 751 11,766
- Zhu , H , Cannon , SB , Young , NB and Cook , DR . 2002 . Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula . Molecular Plant-Microbe Interactions , 15 : 529 – 539 .