ABSTRACT
Apple leaf blotch has been observed in Nelson and Hawke’s Bay regions of New Zealand over recent years. Fungi from the genus Alternaria were consistently obtained from symptomatic leaf material, and morphological and molecular analysis determined that these isolates belong to the Alternaria arborescens complex. Pathogenicity tests were carried out to determine if these fungi were the causal agents of the observed symptoms. Testing was conducted using both orchard and glasshouse grown apple leaves, but symptoms only occurred on young glasshouse grown leaves and only on some of the inoculated leaves. These results confirm that the A. arborescens complex is causing leaf blotch on apple leaves in New Zealand, but also suggest that the fungus is not an aggressive pathogen and disease development is likely dependent on additional factors like leaf age and physiology, climate or growing conditions.
Introduction
Several Alternaria species are known to cause leaf and fruit spots on apple (Malus domestica) (Farr and Rossman Citation2017). The most serious apple pathogen, Alternaria mali, is currently known from America (Canada, Chile, USA), Asia (China, India, Iran, Japan, Korea, Taiwan), Europe (Netherlands, Serbia, Turkey) and Australia (EPPO Citation2016). Due to its potential to cause significant yield loss, this species is listed as an unwanted organism in many apple growing regions, including New Zealand. In addition to A. mali, several other Alternaria species are known to affect apples. For example, recent studies in Australia have isolated a number of apple pathogens belonging to A. alternata, A. arborescens, A. longipes and A. tenuissima species complexes, which can also cause both leaf blotch and fruit spot (Harteveld et al. Citation2013). However, the pathogenicity and virulence of different isolates (both within and between species complexes) were shown to be greatly variable (Harteveld et al. Citation2014).
The only Alternaria species previously known from apple in New Zealand is A. alternata (Farr and Rossman Citation2017). Over recent years, Alternaria isolates different from A. alternata have been consistently isolated from apple leaf lesions in New Zealand, however, these isolates have not been identified to species level (unpublished data). We can now confirm that fungi from the A. arborescens complex are the causal agents of apple leaf blotch in New Zealand with supporting evidence from pathogenicity studies that the detected strains are likely to be weak pathogens.
Materials and methods
‘Envy’ (‘Braeburn’ × ‘Royal Gala’) apple leaves with brown blotch symptoms were submitted to the Plant Health and Environment Laboratory (PHEL) of the Ministry for Primary Industries for disease diagnosis ((A)). Samples were collected from Nelson and Hawke’s Bay regions of New Zealand in early January 2016 and accessioned at PHEL as samples T16_00051 and T16_00052, respectively. Dried leaf material was submitted to the New Zealand Fungarium (PDD) Collection as PDD 108558 (T16_00051) and PDD 108560 (T16_00052).
Figure 1. A, Apple leaf with blotch symptoms submitted to PHEL for disease diagnosis; B, a pure culture of a fungus from Alternaria arborescens complex (ICMP 22004, on potato dextrose agar) obtained from infected apple leaf material.

Leaves were surface sterilised with 70% ethanol and sections of symptomatic leaf tissue were placed on prune extract agar plates (100 ml prune extract, 6 g sucrose, 1.25 g yeast extract and 16 g agar in 900 ml of water). A few symptomatic leaves were also placed in a moist chamber to induce sporulation on plant material. Both the plating assay and moist chamber resulted in consistent and vigorous growth of Alternaria isolates. Subcultures were made from original plates to ensure the isolation of pure cultures. One isolate from each site was used for morphological and molecular identification and submitted to the International Collection of Microorganisms from Plants (ICMP) as ICMP 22004 (T16_00051) and ICMP 22005 (T16_00052).
For morphological identification, the isolates were grown on V8-juice agar media (Mueller et al. Citation2004) in triplicates and incubated with an 8h/16h light/dark cycle at 22°C (following Simmons Citation2007). Colonies were examined for their size, colour and concentric ring development after nine days of incubation. Conidial morphology was examined for shape, colour, size and number of transverse and longitudinal septa using a compound microscope (Leica, Germany).
For molecular identification, DNA was extracted from pure cultures using InviMag® Plant DNA Mini Kit (Invitek, Germany) and Thermo Kingfisher according to manufacturer’s instructions. Molecular identification was completed by amplifying three partial DNA regions: elongation factor (tef1; primers EF1-728F/EF1-986R; (Carbone and Kohn Citation1999); annealing at 58°C for 20 s, 35 cycles), β-tubulin (primers T1/T2; (O’Donnel and Cigelink Citation1997); annealing at 52°C for 55 min, 40 cycles) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; primers gpd1 (Berbee et al. Citation1999)/GPD3r (Inderbitzin et al. Citation2005); annealing at 60°C for 30 s, 40 cycles). Amplified DNA was sequenced by EcoGene (Auckland, New Zealand) and analysed using Geneious 9.1.5 (Biomatters Limited, New Zealand). The sequences of all three gene regions for both isolates were deposited in NCBI GenBank (accession numbers KY965826-KY965831).
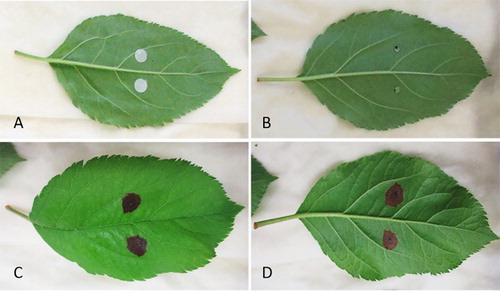
To determine the pathogenicity of the Alternaria isolates, orchard grown young ‘Envy’ and ‘Royal Gala’ leaves and glasshouse-grown young ‘Royal Gala’ leaves were inoculated with a spore suspension, following the general principle of the method described by Harteveld et al. (Citation2014). Two different assays were used: (1) detached leaves placed on trays layered with moist filter paper and enclosed in sealed bags; (2) shoots with attached leaves and a stem end placed in water, enclosed in sealed bags. Four-week-old pure cultures of ICMP 22004 and ICMP 22005 were used as inoculum. Inoculum was introduced either with sterile filter paper discs that were soaked in spore suspension as described by Harteveld et al. (Citation2014) or with 10 µl droplets of spore suspension ((A,B)). To encourage infection, half of the leaves were wounded with a fine sterile needle while the other half were left unwounded. For the control, filter paper discs soaked in sterile water and sterile water droplets were used to inoculate the wounded and unwounded leaves. Inoculated leaves and shoots were incubated for one month in the dark either at 25°C or 30°C.
Figure 2. Pathogenicity assay with A. arborescens (ICMP 22004) on greenhouse grown Royal Gala apple leaves: A and B, freshly inoculated leaves with discs soaked in spore solution (A) and with droplets of spore solution (B); C and D, leaf with leaf blotch symptoms on abaxial (C) and adaxial surface (D).
Results and discussion
The appearance of isolates obtained from infected leaf material was variable and ranged from black to grey with occasional concentric rings formed in the colonies ((B)). Both studied isolates (ICMP 22004 and ICMP 22005) displayed similar morphological features on V8 media, including dark green to black colour colonies with irregular margin and visibly separated 4–6 concentric rings. The colony diameter at nine days after inoculation was 70–80 mm with 5–8 mm white margin. Conidiophores were long and with arborescent appearance. Conidia were ovoid, 12–27 × 6–12 μm, light to medium brown, with 1–4 contrastingly darker transverse septa and no or one longitudinal septum. These morphological characters suggest that these isolates represent Alternaria arborescens (Simmons Citation2007).
Molecular analysis of the fungi isolated from the leaf blotch symptoms determined that the three studied gene regions of the isolates obtained from two different geographic locations were identical. The sequences of the GAPDH region and the β-tubulin region were identical to sequences from a number of A. alternata, A. arborescens and A. tenuissima isolates. This was an expected result as the molecular variation between the small-spored Alternaria species is known to be minimal (eg Woudenberg et al. Citation2015). The tef1 region, however, was identical to A. arborescens isolate CBS 102605 (Genbank accession KC584636), which is the type strain of A. arborescens, and shared only 98–99% identity with sequences from A. alternata and A. tenuissima. A further specific study of the unique fixed nucleotides of the tef1 region confirmed that these isolates belong to A. arborescens complex (sensu Woudenberg et al. Citation2015).
Isolates belonging to A. arborescens complex have been previously reported from apple in Australia and in South Africa (Andrew et al. Citation2009; Harteveld et al. Citation2014). However, in addition to apple, the complex is known to infect a range of different plant hosts and has also been found from non-symptomatic plant material (Woudenberg et al. Citation2015). In New Zealand, isolates from this species complex have been previously isolated from tamarillo, oat and gravel groundsel (Woudenberg et al. Citation2015; Landcare Research Citation2017).
The pathogenicity assay conducted in this study confirmed that the two tested isolates were able to cause leaf blotch on apple ((C,D)). However, only glasshouse grown leaves developed symptoms while no symptoms were produced on inoculated orchard grown leaves. This result indicates that the fungus might only be able to infect apple leaves at certain developmental stage. Moreover, only 25% of glasshouse grown leaves tested had lesions while the majority of infected leaves did not develop any symptoms. The first symptoms were observed around 10 days after inoculation and lesions were evident on leaves incubated at both temperatures (25°C and 30°C), and from leaves inoculated with all different methods (ie filter paper discs and droplet of inoculum; wounded and unwounded leaves; detached leaves and shoots). No disease symptoms were found on control leaves. Sequence-based analysis of the isolates obtained from developed symptoms confirmed that they were identical to the strains that were used to infect the leaves.
Previous studies in Australia have shown that although A. arborescens can be commonly isolated from apple leaf blotch symptoms, there was a great variation in the ability of these fungi to cause plant disease in pathogenicity studies. In a study by Harteveld et al. (Citation2014), among the Alternaria species studied, A. alternata and A. tenuissima caused more severe disease symptoms on apple, and A. arborescens isolates never caused fruit spots. These results from previous and the current studies show that although A. arborescens is commonly found on apple, it is not a significant pathogen and it may not have an economic impact. It is also interesting to note that the favourable environmental conditions that we deployed did not cause consistent development of leaf blotch symptoms on the inoculated leaves. This indicates that the fungi found from apple in New Zealand are not aggressive plant pathogens and the onset of disease is likely influenced by other factors, such as weather and growth conditions in apple orchards.
Acknowledgements
We thank Chris Herries for collecting the original samples from apple orchards, and Peter Wood, Monika Walter, Joy Tyson and Brian Quinn for providing apple leaves and/or shoots for pathogenicity testing. Wellcome Ho is acknowledged for assistance with additional sequence data generation and analysis. We also thank Wellcome Ho, Prasad Doddala, Subuhi Khan and two anonymous reviewers for critical review of the earlier version of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
M. Toome-Heller http://orcid.org/0000-0001-9177-7218
References
- Andrew M, Peever TL, Pryor BM. 2009. An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 101:95–109. doi: 10.3852/08-135
- Berbee ML, Pirseyedi M, Hubbard S. 1999. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 91:964–977. doi: 10.2307/3761627
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556. doi: 10.2307/3761358
- EPPO. 2016. Alternaria mali. EPPO Global Database. [accessed 2016 Dec 6]. https://gd.eppo.int/taxon/ALTEMA.
- Farr DF, Rossman AY. 2017. Fungal databases, Systematic Mycology and Microbiology Laboratory, ARS, USDA. [accessed 2017 Feb 18]. http://nt.ars-grin.gov/fungaldatabases/.
- Harteveld DOC, Akinsanmi OA, Drenth A. 2013. Multiple Alternaria species groups are associated with leaf blotch and fruit spot diseases of apple in Australia. Plant Pathol. 62:289–297. doi: 10.1111/j.1365-3059.2012.02637.x
- Harteveld DOC, Akinsanmi OA, Drenth A. 2014. Pathogenic variation of Alternaria species associated with leaf blotch and fruit spot of apple in Australia. Eur J Plant Pathol. 139:789–799. doi: 10.1007/s10658-014-0433-6
- Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. 2005. Lateral transfer of mating system in Stemphylium. Proc Natl Acad Sci. 102:11390–11395. doi: 10.1073/pnas.0501918102
- Landcare Research. 2017. Systematics collection data. [accessed 2017 April 10]. http://scd.landcareresearch.co.nz.
- Mueller GM, Bills GF, Foster MF. 2004. Biodiversity of fungi: inventory and monitoring methods. Burlington, MA: Elsevier Academic Press.
- O'Donnel K, Cigelink E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus fusarium are nonorthologous. Mol Phylogenet Evolut. 7:103–116. doi: 10.1006/mpev.1996.0376
- Simmons EG. 2007. Alternaria: an identification manual. Utrecht: CBS Fungal Biodiversity Centre.
- Woudenberg JHC, Seidl MF, Groenewald JZ, de Vries M, Stielow JB, Thomma BPHJ, Crous PW. 2015. Alternaria section Alternaria: species, formae speciales or pathotypes. Stud Mycol. 82:1–21. doi: 10.1016/j.simyco.2015.07.001
