Abstract
The groundwater system in Olkiluoto, Finland, is stratified with a mixing layer at a depth of approximately 300 m between sulphate-rich, methane-poor and sulphate-poor, methane-rich groundwaters. New sequence library data obtained by 454 pyrotag sequencing of the v4v6 16S rDNA region indicated that sulphate-reducing bacteria (SRB) dominated the mixing layer while SRB could not be detected in the deep sulphate-poor groundwater samples. With the indispensable support of the sequence data, it could be demonstrated that sulphate was the only component needed to trigger a very large community transition in deep sulphate-poor, methane-rich groundwater from a non-sulphate-reducing community comprising Hydrogenophaga, Pseudomonas, Thiobacillus, Fusibacter, and Lutibacter to a sulphate-reducing community with Desulfobacula, Desulfovibrio, Desufobulbaceae, Desulfobacterium, Desulfosporosinus, and Desulfotignum. Experiments with biofilms and planktonic microorganisms in flow cells under in situ conditions confirmed that adding sulphate to the sulphate-poor groundwater generated growth of cultivable SRB and detectable SRB-related sequences. It was also found that the 16S rDNA diversity of the biofilms was conserved over 103 d and that there was great similarity in diversity between the microorganisms in the biofilms and in the flowing groundwater. This work demonstrates that the presence/absence of only one geochemical parameter, i.e., sulphate, in the groundwater significantly influenced the diversity of the investigated subterranean microbial community.
Introduction
On the island of Olkiluoto, selected for the construction of a deep repository for high-level radioactive wastes, a tunnel denoted ONKALO has been excavated to the future repository area at a depth of 420 m. The current vertical variation in hydrogeochemical groundwater parameters at Olkiluoto comprises a shallow and partly oxygenic freshwater from 0 to approximately 25 m followed by an intermediate-depth brackish, sulphate-rich, methane-poor groundwater with a salinity of less than 1% to a depth of approximately 300 m. In the depth range from 300 m to more than 1000 m, the salinity increases with depth from 1% to >10%, the sulphate concentration is very low or below detection, and the concentrations of hydrogen and methane increase from 1 to >20 μM and 5 mM to >50 mM, respectively. At depths of 250–350 m, there is a layer in which sulphate-rich and methane-rich groundwater mix (Pedersen et al. Citation2008; Posiva Oy 2009).
The construction of the ONKALO tunnel intersects groundwater-conducting aquifers; this generates a drawdown of sulphate-rich groundwater that mixes with the deep methane-rich groundwater. This mixing layer is expected to slowly move deeper in the water-conducting fracture system due to the drawdown effect (Aalto et al. Citation2011). In other words, the construction of ONKALO can be regarded as a large-scale experiment investigating how a slow and continuous mixing of sulphate-rich groundwater with methane-rich groundwater influences microbial diversity and activity.
Initial research into microorganisms in Scandinavian deep granitic aquifers strongly suggested that the vast majority of these microorganisms live attached to surfaces and that they are more metabolically active than are unattached microorganisms (Ekendahl and Pedersen Citation1994; Pedersen and Ekendahl Citation1992a, 1992b). This poses a sampling challenge, because core drilling is required to collect attached microorganisms (Jägevall et al. Citation2011). Due to the obvious risk of the contamination and washout of attached microorganisms by drilling water, an alternative investigation approach is to use in situ experimental installations in underground tunnels (Pedersen Citation2012a) or mines (Lin et al. Citation2006). Flow cells (FCs) incorporating rock surfaces or other solid materials can be installed in contact with deep aquifers under in situ conditions and later be sampled after the attachment of and biofilm formation by groundwater microorganisms.
In this work, deep groundwater from a methane-rich, sulphate-poor aquifer from a depth of 399 m was circulated through crushed rock in FCs for 70 days at in situ pressure. Sulphate was then added to the circulating groundwater, and metabolic activity, numbers of cultivable microorganisms, and 16S rDNA diversity in biofilms and groundwater were studied for 103 d. The effect of adding a 10% portion of groundwater from the mixing layer (318 m), containing a large diversity of sulphate-reducing bacteria (SRB), to methane-rich, sulphate-poor groundwater devoid of SRB was also investigated.
The microbial diversities of cultures were analyzed using cloning and 16S rDNA Sanger sequencing, while the attached and unattached microbial diversities were examined using 454 pyrotag sequencing of the bacterial v4v6 region of 16S rDNA. The concentrations of H2, methane, sulphate, sulphide, ferrous iron, organic acids, and carbon as well as pH and Eh were analyzed. The numbers of cultivable heterotrophic aerobic bacteria (CHAB), SRB, nitrate-reducing bacteria (NRB), iron-reducing bacteria (IRB), autotrophic acetogens (AA), heterotrophic methanogens (HM), and virus-like particles (VLP) as well as the total number of cells (TNC) and amounts of unattached and attached biomass measured as ATP were determined.
Materials and Methods
Groundwater Sources and Characterization
A 76-mm-diameter borehole denoted ONK-KR15 was drilled in the ONKALO tunnel at a depth of 387.9 m (Toropainen Citation2011). The drilling was conducted at an 8.6° inclination on 23–28 February 2011 to a total length of 79.96 m. A metal-free packer system isolated an aquifer in the borehole located 75.0–75.2 m from the tunnel rock face at a depth of 399 m; the transmissivity of the aquifer was 7.6 × 10−9 m2 s−1. The groundwater was directed by this packer system to the FCs described next and then back to the aquifer via two parallel, 1/8-inch polyetheretherketone (PEEK) thermoplastic tubes of high-pressure liquid chromatography quality (IDEX Health and Science, Oak Harbor, WA, USA). The packer system is illustrated in and described in detail elsewhere (Pedersen Citation2005). The system was modified by using a 6-mm stainless steel tube to shield the PEEK tubing in the part of the drillhole exposed to air. The second source of groundwater was a 76-mm-diameter borehole denoted ONK-PVA6 drilled in the ONKALO tunnel at a depth of 318.7 m (Toropainen Citation2009). The drilling was conducted at a 14.8° inclination on 3–4 November 2009 to a total length of 35.15 m. A metal-free packer system isolated an aquifer in the borehole located 32.7–32.9 m from the tunnel rock face at a depth of 327 m. Groundwater samples for chemical analysis were collected from these boreholes on 12 April 2012 and immediately transported to Teollisuuden Voima, where the chemical analyses were performed according to internal protocols or were subcontracted to external laboratories as described in detail elsewhere (Supplementary in Pedersen et al. Citation2008).
Table 1. Geochemical parameters of the experimental groundwater and a comparative ratio for each parameter
Table 2. Concentrations of dissolved gases in ONK-KR15 groundwater, sampled on 11 January 2012, in ONK-KR15 groundwater used to fill the flow cell circulation systems sampled on 17 April 2012 and in ONK-PVA6 groundwater
Table 3. The most probable numbers of cultivable microorganisms and the total number of cells in groundwater from borehole ONK-KR15 sampled on 11 January 2012 and from borehole ONK-PVA6 sampled on 17 April 2012; SD = standard deviation, n = number of observations
Fig. 1. Images of the experimental systems. (a) The packer system used to isolate the aquifer to which flow cells (FCs) with crushed rock were connected. (b) The FCs were installed underground in series, four by four, in racks with flow meters and pumps. Groundwater was circulated from the isolated aquifer through the FCs and back to the aquifer for 70 days. The FCs were then disconnected and transferred to (c) temperature-controlled flow cell circulation systems in the laboratory.
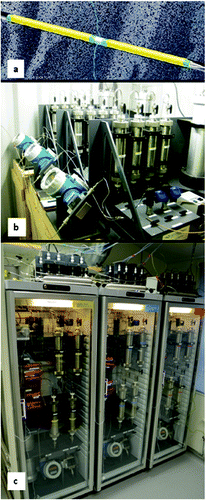
FC Systems for Field Work
Three identical FC field systems comprising four FCs each, a micropump (Micropump GAH, series V21 J with a PEEK impeller; Labinett, Göteborg, Sweden), two pressure meters (S-11, 40 Bar 4–20 G1/2; WIKA – AB Svenska Industri Instrument, Göteborg, Sweden), a flow meter (Promag 50; Endress+Hauser Flowtech AG, Sollentuna, Sweden), and a 4-L expansion vessel (Pedersen Citation2005) were installed in a container placed in the ONKALO tunnel at a depth of 387 m and connected to the packer system in ONK-KR15 (). Each FC consisted of a steel tube (length 300 mm, diameter 65 mm) lined with polyvinyldifluoride (PVDF) plastic. Each FC also had a 120-mm-long PVDF insert with a 22 × 32-mm opening that supported 110 g of crushed rock grains offering a rock surface area of approximately 850 cm2 per FC for microbial adhesion and biofilm formation, assuming spherical rock grains with an average diameter of 3 mm.
The rock grains, which were heat sterilized (160°C for 5 h), were obtained from the drill core of the ONK-KR15 drillhole at the approximate location of the intersected aquifer. Three flow stabilizers at each end of the insert ensured an evenly distributed slow laminar flow of water through each FC (Pedersen Citation1982). The FCs were installed on 7 February 2012. Groundwater was circulated through them at an in situ pressure of 3.2 MPa for 70 d at a flow rate from and to the aquifer of 22–25 mL min−1. The total volumes of groundwater circulated were 2562, 2381, and 2287 L in the three field systems, respectively, as registered by the flow meters.
Configuration of Growth Experiments
The 12 FCs exposed to ONK-KR15 groundwater for 70 d were transported under pressure from the ONKALO tunnel to the laboratory in Mölnlycke, Sweden, and four replicate FCs were installed in each of three flow cell circulation systems (FCCSs), resulting in a total of three treatment possibilities (). Sulphate and ONK-PVA6 groundwater were then added as follows: Three Teflon-lined, 500-mL stainless steel cylinders (304L-HDF4-500-T; Swagelok, Göteborg, Sweden) were filled at room temperature (RT, 20°C) with: 1) 500 mL of ONK-KR15 groundwater, 2) 5 mmol Na2SO4 dissolved in 500 mL of ONK-KR15 groundwater, and 3) 5 mmol Na2SO4 dissolved in 500 mL of ONK-PVA6 groundwater. Each cylinder was connected in line with the circulating groundwater in one FCCS, resulting in a total circulating volume of 5500 mL per FCCS.
These treatments are hereafter denoted control, sulphate, and sulphate + ONK-PVA6. The start date for these groundwater circulations was 26 April 2012 and the end date was 7 August 2012, the duration of the experiment being 103 d. The flow rate was kept at 22–25 mL min−1, corresponding to a flow of approximately 1 mm s−1 over the rock grains. Four pressure-resistant microsensor Eh electrode couples equipped with one platinum micro-electrode with a tip diameter of 400–600 μm (RD500; Unisense A/S, Aarhus, Denmark) and one Ag/AgCl reference electrode with a tip diameter of 90–110 μm in gel-stabilized electrolyte (REF100; Unisense) were installed in line in each FCCS. The electrodes represented an adaptation of the standard glass Unisense microsensors mounted in the stainless steel flow cells. The electrodes were connected to two eight-channel mV amplifiers that transformed the recorded voltages into digital signals, which were subsequently collected and stored in Microsoft Office Excel files every 600 s using SensorTrace Basic software (version 1.9; Unisense A/S).
Complete sampling was performed six times, i.e., on days 0, 7, 19, 40, 61, 82, and 103, for analysis as described next. On each sampling occasion, 20 mL of circulating water was drained and discharged; two 25-mL volumes of water were collected in sterile 50-mL polypropylene (PP) tubes (Sarstedt, Landskrona, Sweden) and deep frozen until sulphate analysis, and 10 mL of water was collected in a sterile 15-mL PP tube for immediate ATP analysis. Six 10-mL volumes of water were collected, using syringes, in butyl rubber-stoppered anaerobic glass tubes (no. 2048-00150; Bellco Glass, Vineland, NJ, USA) for MPN analysis and 10 mL was collected for CHAB analysis. Two 10-mL volumes of water were collected in PP tubes, preserved with 0.02 μm of filtered, neutralized formaldehyde to a final concentration of 2.5%, and analyzed for TNC and VLP, respectively. Thereafter, 9 mL of water was sampled for sulphide analysis, and two 5-mL volumes were sampled using a 0.2-μm syringe filter (Minisart, Sartorius syringe filter, hydrophilic; Fisher Scientific, Göteborg, Sweden) and stored at −20°C until acetate and lactate analyses could be performed.
Next, 25 mL of water was sampled using a 0.2-μm syringe filter (Minisart) for immediate ferrous iron analysis. Two 10-mL volumes of water were sampled using a 0.2-μm syringe filter (Minisart) and deep frozen until DOC analysis. Finally, 10 mL of groundwater was collected for pH analysis and 100 mL for gas analysis. In total, 334 mL of water was sampled on each sampling occasion. After sampling the water on days 0 and 103, one batch of rock grains was collected from each of two FCs in each FCCS for subsequent analysis of the amount of attached ATP and the 16S rDNA diversity.
Acetate, Lactate, Organic Carbon, Ferrous Iron, Sulphate and Sulphide Analysis and pH
Acetate and lactate concentrations were determined using the enzymatic UV method (kit no. 10148261035 for acetate and kit no. 10139084035, for lactate; Boehringer Mannheim/R-Biopharm AG, Darmstadt, Germany) using a Genesys 10UV spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) for detection. Samples for dissolved organic carbon (DOC) analysis were diluted 1–100 times before analysis to obtain the optimal analytical concentration range. Samples of 25 mL were filtered through 0.2-μm hydrophilic syringe filters (Minisart) and deep frozen at −20°C until analysis at ALS Scandinavia AB (Täby, Sweden) according to the CSN EN 1484 method. The uncertainty was ±20% of the analyzed values. Sulphate was analysed using the SulfaVer 4 method (method no. 8051, programme 680; HACH Lange AB; range 0.03–0.73mM with 95% confidence limits of distributionof ±10%). Sulphide was analyzed using a colorimetric methylene blue method with an uncertainty of ±17% (Swedish Standard Method SIS 028115). Ferrous iron concentrations were determined using the 1-10 phenanthroline method (method no. 8146, program 255, range 0.4–54 mM with 95% confidence limits of distribution of ±11%; HACH Lange AB, Stockholm, Sweden). The pH of 5-mL subsamples was determined immediately following extraction from the FCCSs, using a Schott CG84310 pH meter (Schott AG, Mainz, Germany) fitted with a BlueLine 13 pH electrode (VWR, Stockholm, Sweden) calibrated according to the manufacturer's instructions.
ATP Analysis
The ATP Biomass Kit HS (no. 266–311; BioThema, Handen, Stockholm) was used to determine total ATP in cells living in groundwater. The ATP biomass method used here has been described, tested in detail, and evaluated for use with Fennoscandian Shield groundwater (Eydal and Pedersen Citation2007). The method was also used for biomass attached to the rock grains, but with the following modification: Approximately 10 rock grains were sampled from each of two FCs per FCCS and placed in ATP extraction solution and analyzed.
TNC and VLP
The TNC mL−1 was determined in 10-mL samples using the acridine orange direct count method as devised by Hobbie et al. (Citation1977) and modified by Pedersen and Ekendahl (Citation1990). The total number of VLP was determined using a direct count method with SYBR Gold (Molecular Probes, Eugene, OR, USA) according to Noble and Fuhrman (Citation1998).
Gas Sampling and Analysis
Water samples were collected using a pressure vessel as described elsewhere (Hallbeck and Pedersen Citation2008). The sample was transferred to a vacuum container and any gas in the water was boiled off under vacuum (i.e., water vapor pressure) at RT; the transfer time was approximately 20–30 min. After extraction, the gas was compressed and transferred to a 10-mL syringe (SGE Analytical Science, Melbourne, Victoria, Australia) and the volumes of extracted gas and water were measured. The captured gas was subsequently transferred to a 6.6-mL glass vial stoppered with a butyl rubber stopper and sealed with an aluminium crimp seal. The vial had previously been evacuated and flushed twice with N2, and left under high vacuum (1 Pa). A silica gel dehydrant was added to adsorb any traces of water remaining in the gas. Analysis was then performed using gas chromatography.
Two different chromatographs were used and equipped as follows. H2 (<20 ppm) Ar and CO2 were analyzed on a Bruker 450 gas chromatograph equipped with a CP7355 PoraBOND Q 50 m × 0.53 mm ID column, a CP7536 MOLSIEVE 5A PLOT 25 m × 0.32 mm ID column, and a pulsed discharge helium ionization detector (PDHID) (Bruker Daltonics Scandinavia AB, Solna, Sweden). He and N2 were analyzed on a Varian Star 3400CX gas chromatograph (Varian Analytical Instruments, Varian AB, Bromma, Sweden) using a thermal conductivity detector with oven, detector, and filament temperatures of 65, 120, and 250°C, respectively. The gases were separated using a Porapak-Q column (2 m × 1/8 inch diameter; Sigma-Aldrich, St. Louis, MO, USA) followed by a molecular sieve 5A column (6 m × 1/8 inch; Sigma-Aldrich) with argon as the carrier gas. CH4, C2H6, and CO were analyzed on a Varian Star 3400CX gas chromatograph (Varian Analytical Instruments) using a flame ionization detector (FID) with an oven temperature of 65°C and a detector temperature of 200°C. The gas was separated using a Porapak-Q column (2 m × 1/8 inch diameter; Sigma-Aldrich) and analyzed on the FID with N2 as the carrier gas.
Cultivation Media
Media were prepared for CHAB and for the most probable number analysis of NRB, IRB, MRB, SRB, AA, and AM as described elsewhere (Hallbeck and Pedersen Citation2008). The cultivation time was about 8 weeks to ensure that slow-growing microorganisms would be included in the results.
DNA Extraction from MPN Cultures, Groundwater, and Biofilms
Total genomic DNA from groundwater and most probable number (MPN) cultures was extracted according to the manufacturer's protocol using the MO BIO PowerWater DNA isolation kit (cat. no. 12888) and from biomass attached to rock grains using the MO BIO PowerBiofilm DNA isolation kit (cat. no. 24000–50), both from MO BIO Laboratories, Carlsbad, CA, USA. The extraction volume of the MO BIO extraction kits used in this work was 100 μL.
Groundwater was pressure filtered using high-pressure, stainless steel, 47-mm filter holders (X4504700; Millipore AB, Solna, Sweden) equipped with the water filters from PowerWater kit filter units (MO BIO Laboratories). The filter holder was equipped with a pressure relief valve (Swagelok SS-RL3S6MM; SWAFAB, Sollentuna, Sweden) and a manometer that enabled adjustment of a pressure drop over the filter between 200 and 400 kPa relative to the ambient aquifer pressure. Groundwater was filtered at a flow rate of 0.05–0.2 L min−1 from the ONK-PVA6 and ONK-KR15 tunnel boreholes on 17 April 2012. The approximate filtered volumes of groundwater were 57 L for the high-permeability aquifer in ONK-KR15 and 14 L for the low-permeability aquifer in ONK-PVA6.
From selected MPN cultures, 9 mL of culture was filtered onto 0.22-μm water filters (cat. no. 14880-100-WF; MO BIO Laboratories) using vacuum suction. Using sterile, DNA-free tweezers, 40 rock grains were collected from each of the three FCCSs and pooled (2 g of rock) on day 0, and 80 rock grains were collected from each FCCS and pooled (4 g of rock) on day 103; these rock grains were placed directly into empty biofilm DNA extraction vessels provided by the manufacturer.
Total extracted nucleotide concentrations were measured using the ND-1000 UV-vis spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA) and double-stranded (ds) DNA concentrations were measured fluorometrically using the Stratagene MX3005p fluorometer with MXPro software (Agilent Technologies, Santa Clara, CA, USA) and the Quant-it Picogreen reagent kit (cat. no. P7589; Molecular Probes), according to the manufacturer's specifications. The extracted DNA was stored at −20°C and subsequently used for sequencing.
Cloning and 16S rDNA Sequencing of MPN Cultures
The species diversity of MPN culture microorganisms was gauged by their bacterial 16S rDNA sequence. The PCR amplification used the universal 16S rDNA forward and reverse primers 27f and 1492r, respectively (Lane Citation1991). Thermal cycling conditions were 98°C for 30 s, 30 cycles of 98°C for 30 s, 60°C for 30 s, and 72°C for 30 s with a final extension of 72°C for 5 min. The amplification products were visualized using gel electrophoresis on 1% agarose gel, stained with ethidium bromide, and illuminated by UV exposure. Amplification products were then purified using a QIAquick Gel Extraction Kit (cat. no. 28704; QIAGEN, Solna, Sweden) following the manufacturer's protocol. To produce 3´A overhangs of the blunt-ended iProof polymerase product, 1 μL of Taq polymerase (cat. no. 18038–042; Molecular Probes) and additional dATP were added to the reaction mixtures, which were then incubated for 30 min at 72°C.
The purified samples were cloned into the linearized PCR 2.1-TOPO vector and transformed into chemically compete-nt TOP10´F Escherichia coli cells using the TOPO TA cloning kit (cat. no. K4550-01; Molecular Probes) following the manufacturer's protocol. White clones containing the insert were randomly selected and each colony was inoculated on-to LB agar plates containing kanamycin (40 mg mL−1) and incubated overnight at 37°C. The recombinant plasmids were extracted and subsequently sequenced using the Value Read Plate service (Eurofins MWG Operon, Ebersberg, Germany) with the M13rev(−29) sequencing primer (5´-CAGGAAACAGCTATGAC-3´) and the M13uni(−21) sequencing primer (5´-TGT AAAACGACGGCCAGT-3´) provided by Eurofins MWG for the Value Read Plate service for the 16S rDNA clones.
Raw data sequences were screened for chimeric sequences using the Bellerophon program (Huber et al. Citation2004). Sequence data were analyzed and aligned using the Geneious 6.0.3 software package (Biomatters, Auckland, New Zealand). The 16S rDNA reference gene E. coli Brosius with accession number J01695 was used as a sequence mask for aligning the 16S rDNA clones (Huber et al. Citation2004). In addition, the clones were compared with sequences available in the BLAST nucleotide database. Sequence homology was analyzed using either the nucleotide–nucleotide algorithm or the 16S rDNA microbial algorithm. Sequences that were <99.9% similar to database records were submitted to the GenBank database under accession numbers KC676781 to KC676786.
454 Pyrotag Sequencing, Processing, and Analysis of DNA from Groundwater and FC Biofilms
The degenerate forward 518F (5´-CCAGCAGCYGCGGTAA-3´) and reverse 1064R (5´-CGACRRCCATGCANCACCT-3´) primers targeting the v4v6 region of the bacterial 16S rDNA were used for pyrotag sequencing on a 454 Roche GS-FLX system (454 Life Sciences, Branford, CT, USA) using the Roche Titanium protocol for generating reads as part of the Census of Deep Life initiative (http://www.deepcarbon.net/content/deep-life). The beginning and end of each read were trimmed for primer bases, and sequences likely to be of low quality based on assessment of pyrotag sequencing error rates were removed (Huse et al. Citation2007). The 454 pyrotag sequence processing to assign a taxonomic classification was done using the Global Alignment for Sequence Taxonomy (GAST) tag mapping methodology (Sogin et al. Citation2006), in which the reference database of 16S rDNA, RefSSU, was based on the SILVA database (Pruesse et al. Citation2007). If two-thirds or more of the full-length sequences shared the same assigned operational taxonomic unit (OTU), the tag was assigned to that OTU. Tags that did not match any reference tag according to BLAST were not given a taxonomic assignment.
Further details of methodology and library construction can be found elsewhere (Marteinsson et al. Citation2013). The representativeness of sequences was tested by rarefaction analysis and the Chao index was used to estimate OTU richness. To statistically estimate the abundance and evenness of each sample, Shannon and Simpson indices were calculated. Distance calculations for sequence similarities were performed using the Morsita–Horn algorithm. The data generated have been submitted to the NCBI Sequence Read Archive (SRA) with accession numbers SRX268395 and SRX268398–SRX268402. Sequences appearing at approximately 3% or more frequency-abundance were searched using BLAST against the GenBank nucleotide database and sample sites of the closest match were registered. These sequences were aligned using BioEdit 7.1.3.0 (Tom Hall, Ibis Biosciences, Carlsbad, CA, 92008) and the identity of sequences between samples was analyzed.
Bioinformatics and Statistical Analyses
The 454 data were evaluated using the Visualization and Analysis of Microbial Population Structure (VAMPS) website (www.vamps.ml.edu). Data graphics design and statistical analyses were performed in Statistica 10 (Statsoft, Tulsa, OK, USA).
Results
Groundwater Characterization
The geochemistry of groundwater from ONK-PVA6 and ONK-KR15 was analyzed on 12 April 2012 (). The ratios of the groundwater parameters shown in indicate that the two experimental groundwater types only differed substantially in their content of sulphur compounds; otherwise, most other parameters differed at most by a factor of 2, which can mainly be ascribed to the higher salinity of ONK-KR15 than of ONK-PVA6 groundwater. The composition of dissolved gases was analyzed twice in ONK-KR15 groundwater, on 11 January and 17 April 2012, and once in ONK-PVA6 groundwater, on 18 September 2012 (). More dissolved gas, particularly methane and hydrogen, was contained in ONK-KR15 than in ONK-PVA6 groundwater.
There were 1.8 × 104 cells mL−1 and 4.5 × 103 amol ATP mL−1 in the groundwater from OL-KR15 sampled on 11 January 2012 (). NRB dominated the MPN determination, with only the MPN of NRB and MRB being above the detection limit. There were 4.9 × 104 cells mL−1 and 1.06 × 104 amol ATP mL−1 in the groundwater from OL-PVA6 sampled on 17 April 2012 (). IRB and MRB dominated the MPN determination and the MPN of NRB and SRB were also above the detection limit.
TNC, VLP, and ATP in the FCCSs
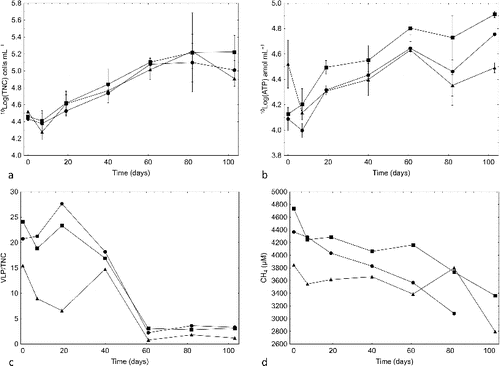
The total number of cells mL−1 and the amount of ATP mL−1 did not differ significantly between the FCCSs for most of the experimental time, but there was an increasing trend in all three systems ( and ). Similarly, the amounts of ATP g−1 of rock grains differed significantly over time only in the sulphate + ONK-PVA6 FCCS (). The numbers of VLPs were approximately constant in all FCCSs and the numbers of VLPs per cell were highest in the control and the sulphate FCCSs, while this number was lower in the sulphate + ONK-PVA6 FCCS (). The number of VLPs per cell decreased significantly after approximately 40 d from an average of 20 to an average of 2–3 for the remaining experimental time. This decrease in VLPs per cell was correlated with increasing TNC and ATP concentration. The lytic activity of phages appears to have decreased over time, allowing room for an increase in unattached biomass.
Table 4. Amount of ATP on rock grains from FC 2 (n = 3) and FC 4 (n = 3) in each flow cell circulation system
Fig. 2. (a) Total number of cells (TNC), (b) ATP concentration, (c) number of virus-like particles (VLPs) per total number of cells (TNC), and (d) concentration of dissolved methane in groundwater circulating through the three flow cell cabinets supplemented with 500 mL of ONK-KR15 groundwater (•), 500 mL of ONK-KR15 groundwater and 1 mM NaSO4 (▪), and 500 mL of ONK-PVA6 groundwater and 1 mM NaSO4 (▴). Bars indicate ±1 standard deviation; n = 3 in (a)–(b).
Cultivated Microorganisms and Organic Acids in the FCCSs
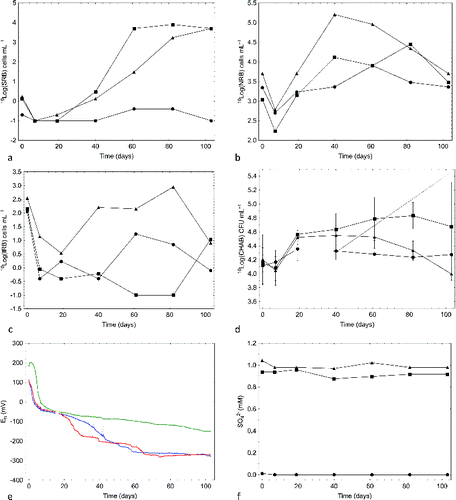
The MPN of SRB increased to approximately 104 cells mL−1 in both sulphate-amended FCCSs and was around the detection limit (i.e., 0.2 cells mL−1) in the sulphate-poor control FCCS (). The MPN of NRB was highest in the sulphate + ONK-PVA6 FCCS after 40 d (). At the start and the end of the experiment, all three FCCSs had a similar MPN of NRB. The MPN of IRB varied over time and was highest in the sulphate + ONK-PVA6 FCCS (). Likewise, the numbers of CHAB were approximately similar throughout the experiment in all FCCSs, averaging approximately 5 × 104 cells mL−1 (). The MPN of AA was at the detection limit in the control system and increased to at most 1–10 AA mL−1 in the sulphate-amended system, and acetate was not produced in any of the FCCSs. The MPNs of HM were below detection limits on all sampling occasions in all FCCSs. The concentrations of acetate and DOC did not change relative to the starting values on sampling occasion 0 of 18 μM acetate and 0.25 μM DOC, respectively.
Fig. 3. (a) Most probable number (MPN) of sulphate-reducing bacteria (SRB), (b) MPN of nitrate-reducing bacteria (NRB), (c) MPN of iron-reducing bacteria (IRB), (d) cultivable heterotrophic aerobic bacteria (CHAB), (e) Eh measured using internal electrode couples: average of four electrode signals (blue line in e), in the sulphate flow cell circulation system (FCCS) (red line in e), and in the sulphate + ONK-PVA6 FCCS (green line in e), and (f) sulphate concentration in groundwater circulating through the three FCCSs supplemented with 500 mL of ONK-KR15 groundwater (•), 500 mL of ONK-KR15 groundwater and 1 mM NaSO4 (▪), and 500 mL of ONK-PVA6 groundwater and 1 mM NaSO4 (▴).
Chemistry in the FCCSs
The methane concentrations decreased by approximately 15% relative to the starting values (). There was less methane in the sulphate + ONK-PVA6 FCCS because the added ONK-PVA6 groundwater contained less methane than did ONK-KR15 groundwater (). The pH decreased at roughly the same rate in all FCCSs, declining from a starting value of approximately 8.2 to approximately 7.5 after 103 d. The Eh of the control and the sulphate FCCSs decreased to a steady level of approximately −250 mV after 70 d, as registered by the internal microelectrodes (). The Eh of the sulphate + ONK-PVA6 FCCS slowly decreased to approximately −100 mV by the end of the experiment. The sulphate additions resulted in 1 mM sulphate in the sulphate FCCS and in a somewhat higher concentration in the sulphate + ONK-PVA6 FCCS; this difference was due to a high sulphate concentration in the added ONK-PVA6 groundwater (1.9 mM) (). The sulphate concentration did not change over the experimental time and was below detection in the control FCCS. The sulphide concentration was below detection in all FCCSs on all sampling occasions. The ferrous iron concentration increased to approximately 20 μM in all FCCSs.
Cloning and 16S rDNA Sequencing of MPN Cultures
DNA extracted from the highest dilutions of positive NRB and SRB cultures inoculated on days 82 and 103 were cloned and sequenced (). The sequence data revealed a total of eight clone taxa from the studied MPN cultures. The sequences found represented the Deltaproteobacteria, Alphaproteobacteria, and Gammaproteobacteria phyla. The dominant species affiliations were to Desulfovibrio aespoeensis and Pseudomonas stutzeri with 41 and 14 clone observations, respectively; these clones were 99.9–100% similar to database records in NCBI GenBank.
Table 5. Taxa detected in clones from MPN cultures of SRB and NRB sampled on days 82 and 103 from the sulphate and the sulphate + ONK-PVA6 flow cell circulation systems
DNA Recovery in the Groundwater and Biofilm Extractions
The total amounts of extracted DNA ranged from 26 to 243 × 10−9 g (). The average amount of DNA in a typical groundwater bacterium, for example, D. aespoeensis (Motamedi and Pedersen Citation1998), is 649 Daltons/base pair × 3,629,109 bases (Locus CP002431) = 2.36 × 109 Daltons cell−1 = 2.36 × 109 Daltons cell−1 × 1.6605402 × 10−24 g Dalton−1 = 3.9 × 10−15 g DNA cell−1. Approximately 57 L of groundwater was filtered from ONK-KR15 and the DNA recovery was 63 × 10−9 g of DNA (), which corresponds to 1.62 × 107 average-sized cells based on the DNA estimate for D. aespoeensis. There were 1.8 × 104 cells mL−1 in ONK-KR15 groundwater at the start of filtration ().
Table 6. Amounts of extracted double-stranded DNA analysed fluorometrically using the Stratagene MX3005p fluorometer with MXPro software and the Quant-it Picogreen reagent kit from Molecular Probes; observed and estimated diversity at total OTU level (>0% sequence abundance) in groundwater and biofilm sequence libraries
The calculated average DNA recovery then becomes 1.6%. Calculated in the same way, the DNA recovery was 9.2% for ONK-PVA6 groundwater. The number of cells in deep groundwater tends to decrease during aquifer drainage; if this occurred here, the DNA recovery was larger because a decreasing TNC during filtration will increase the observed amount of DNA over the total number of cells captured on the filter. Using the same formula for biofilms suggests that there were approximately 2 × 106 cells (g rock grains)−1, which is more than the ATP analysis indicated, i.e., approximately 5 × 105 cells (g rock grains)−1, assuming an average of 0.4 amol ATP per cell (Eydal and Pedersen Citation2007). It is impossible to decide whether ATP underestimated the biomass or whether free dsDNA was adsorbed on the rock from cells lysed by phages or from dead adsorbed cells. The three different assays, i.e., DNA, TNC, and ATP, agreed reasonably well, given the relatively large uncertainties with the input data for these calculations.
Groundwater 16S rDNA v6v4 Sequence Diversity
Except for 11 Desulfosporosinus reads ( = 0.06%), sequences related to SRB were absent in the sequence library from ONK-KR15 groundwater (). Not even singletons of other SRB OTUs were found among the 18134 reads. Rarefaction curves indicated that 90–95% of the 16S rDNA diversity was captured for each sample. This observation agrees well with the lack of cultivable SRB (). The dominant sequences were closely related to the hydrogen-oxidizing genus Hydrogenophaga (30.3%) (Willems et al. Citation1989) followed by Pseudomonas- (8.8%) and Thiobacillus- (8.6%) related sequences ().
Table 7. Occurrence of sulphate-reducing taxa (Deltaproteobacteria and Firmicutes) in sequence libraries from biofilms and groundwater at ≥0.1% abundance and observed major OTUs in the libraries and their reported sample sites in the GenBank nucleic acid database
Fig. 4. Composition of v4v6 pyrotag sequencing libraries for samples of ONKALO groundwater and FCCS biofilms. Sequences with ≥1% frequency-abundance are shown. Bar designations: KR15 = ONK-KR15 groundwater sampled on 17 April 2012; bf 0 = biofilm on day 0; bf C = control biofilm on day 103; bf S = sulphate biofilm on day 103; bf S 6 = sulphate + ONK-PVA6 biofilm on day 103; PVA6 = ONK-PVA6 groundwater sampled on 17 April 2012. NA: not annotated. The tree above the bar graph depicts a Morisita–Horn distance measure, constructed using an unweighted pair group method with arithmetic mean (UPGMA) with taxonomic depth at the species level. The scale bar represents 5% nucleotide substitutions.
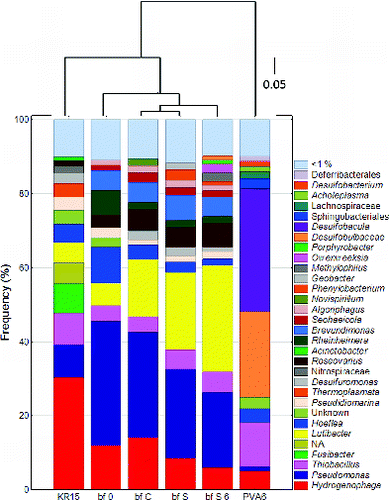
Archaea were represented by sequences related to Euryarchaeota and to Thermoplasmata (3.5%), the latter sequence having previously been described in samples from deep South African goldmines (Gihring et al. Citation2006). Unlike the sequence library from ONK-KR15 groundwater, that from ONK-PVA6 groundwater was dominated by 454 sequences related to genus or family of SRB, represented by Desulfobacula (33.3%) and Desulfobulbaceae (23.2%), respectively (). The three most abundant sequences from ONK-KR15 were also found to some extent in the ONK-PVA6 sequence library. The incidence of similar sequences in the ONK-PVA6 and ONK-KR15 sequence libraries is reasonable, because hydrological modelling suggests that intermediate-depth, sulphate-rich groundwater containing SRB penetrates downwards and mixes with the deep, sulphate-poor groundwater in the aquifer region to which ONK-PVA6 is connected (Aalto et al. Citation2011).
Biofilm 16S rDNA v6v4 Sequence Diversity
A clear phylogenetic similarity was observed among biofilm samples, and the diversity profile of each FCCS treatment indicated little change over 103 d relative to the biofilm diversity on day 0 (). Many of the OTUs found in ONK-KR15 groundwater were also found in the FC biofilms. However, some genera seemed to prefer the planktonic state as they were not found, or found at very low frequencies, in the biofilms. Sequences related to the Fusibacter, Thermoplasmata, and Nitrospira OTUs were not found in the biofilm sequence libraries. Others, such as the Brevundimonas OTU, were ten times more abundant in the biofilms than in the groundwater libraries.
Otherwise, most OTUs found in the groundwater were also represented in a generally comparable order of frequency-abundance in all biofilm samples. The Hydrogenophaga, Lutibacter, and Pseudomonas OTUs together constituted most of the sequence reads in all biofilm samples at >50% sequence abundance and constituted 45% of the sequence reads in ONK-KR15 groundwater. The biofilm diversity appeared to constitute a good predictor of groundwater diversity and vice versa.
The MPN of SRB in the sulphate-amended FCCS was 5000 cells mL−1 on day 103 (), indicated by cloni-ng to be D. aespoeensis (), while the control was below detection (<0.2 cells mL−1). The sequence data agreed with this, with only 11 reads related to Desulfobulbaceae in the contr-ol biofilm library on day 103, yet the sulphate biofilm library had a total of 218 reads related to SRB, of which 168 (0.81%) were related to D. aespoeensis. The sulphate + ONKPVA6 biofilm library similarly contained 211 reads related to SRB with 43 related to D. aespoeensis and 22 related to the Desulfobacula OTU. Details on the frequency-abundance of SRB are presented in .
Discussion
As outlined in the Introduction, two major hydrogeochemical types of groundwater are layered over depth in Olkiluoto separated by a mixing layer between 250 and 350 m in depth. Consequently, this variation over depth presents distinctly different geochemical environments for microbial diversity and activity. The 454 sequence diversities of these layers differed considerably, with most SRB-related sequences in the ONK-PVA6 groundwater library representing the mixing layer (). This agreed with the results for cultivable microorganisms () and with previous observations of a significant reduction of sulphate to sulphide in the mixing layer (Pedersen et al. Citation2008). In contrast, the deep groundwater system that feeds ONK-KR15 appeared “desert-like” with a very low diversity of cultivable cells, i.e., only facultative anaerobic bacteria with a nitrate-reducing capacity ().
Notably, the absence of sulphate appeared to exclude SRB-related sequences from the ONK-KR15 groundwater and biofilm sequence libraries. However, the species richness (i.e., number of taxons >0%) and the calculated sequence diversity indexes () were somewhat greater for ONK-KR15 than for ONK-PVA6 groundwater. The most abundant sequences in the ONK-KR15 sequence library were related to the potentially autotrophic, hydrogen-utilizing genus Hydrogenophaga (Willems et al. Citation1989) and to Pseudomonas, Thiobacillus, and Fusibacter (Ravot et al. Citation1999). The media used for cultivation were not supplemented with the electron donors and acceptors used by these genera, such as hydrogen and thiosulphate, which might have favoured their growth. In view of these new results, cultivation media can now be redesigned to increase the variety of cultivable microorganisms. Because PCR primers specific to Archaea were not used in this analysis, the task of analyzing the full Archaeal diversity in ONKALO groundwater remains.
Observed Sequence Diversity
Over the 103-d experimental time and the two treatments, the diversity of the biofilms changed only slightly (). Alignment analysis of OTU sequences appearing in abundances >3% indicated that they were identical between the groundwater and all four biofilm samples, except for the Hydrogenophaga sequences, which varied somewhat in OTU composition between the samples. Similarly, biomass analyzed as ATP () and biofilm diversity changed little over 103 d, implying a constant level of diversity in the ONK-KR15 biofilms. The reproducible 454 pyrotag sequencing results for biofilms over treatments and time clearly attest to the reproducibility and robustness of the DNA extraction and 454 pyrotag sequencing methodology, in which each biofilm can be regarded as an independent sample for sequencing.
The numbers of unique OTUs observed were in the 108–135 range (), more than previously found in Kalahari (Gihring et al. Citation2006) and Fennoscandian Shield (Pedersen et al. Citation1996, 1997) groundwaters using cloning and sequencing, but somewhat fewer than found using 454 pyrotag sequencing on subglacial waters (i.e., 208-410 OTUs; Marteinsson et al. Citation2013) and over 10 times fewer than found in deep seawater (Sogin et al. Citation2006). The observed 454 sequences were generally comparable to sequences from sediments, salt marshes, groundwater, sludge, and lake waters reported to GenBank (). However, although the similarities were close to 100%, the Blast results were too diverse to permit conclusions as to the global site specificity of the observed OTUs from ONKALO.
The deep saline and slightly alkaline groundwater of Olkiluoto may be under the influence of a much deeper ultramafic environment with ongoing serpentinization, which could explain the observed very high concentrations of methane and hydrogen (Sherwood Lollar et al. Citation1993). Such subterranean environments seem to enrich Hydrogenophaga elsewhere (Brazelton et al. Citation2013). A similar enrichment was suggested in the case of ONK-KR15 because this genus dominated the 454 sequence library. The few percent of Archaea sequences in the data set were related to Thermoplasmata sequences from deep, saline groundwater from South African goldmines (Gihring et al. Citation2006; Takai et al. Citation2001). The domination of Hydrogenophaga OTUs indicates that the diversity of deep subterranean life is controlled by environmental parameters that regulate the growth and activity of various genera with relevant physiological and metabolic capacities. Major relevant parameters would be the presence and absence of various carbon and electron donors and electron acceptors.
Effect of Sulphate on SRB Diversity and Activity
The only major difference in geochemistry between ONK-KR15 and ONK-PVA6 groundwaters was the absence of sulphate and sulphide from ONK-KR15 groundwater (). This difference profoundly influenced the observed numbers of cultivable SRB and the 16S rDNA diversity of SRB, which were great in ONK-PVA6 groundwater and virtually absent from ONK-KR15 (, and ). With sulphate being the main available electron acceptor in the mixing layer, methane could potentially be the largest electron donor for sulphate reduction, 10 times larger in concentration than the second-largest potential electron donor, DOC ( and ). However, unlike methane, which, if oxidized by microorganisms, can be continuously replenished from the deep methane-rich layer via diffusion and groundwater movement, DOC must be synthesized by chemo- or photo-autotrophic metabolic processes.
In a previous study, the production of at most 320 μM acetate was observed in FCCSs charged with ONK-PVA6 groundwater, and it was speculated that the acetate was produced by microorganisms using methane as the source of carbon (Pedersen Citation2013). Several of the SRB in ONK-PVA6 () are known as complete acetate-oxidizing genera, i.e., Desulfobacterium (Brysch et al. Citation1987), Desulfobacula, and Desulfotignum (Kuever et al. Citation2001). Consequently, in agreement with the presence of acetate and its possible production from methane, present and previous results suggest that the mixing of intermediate-depth sulphate-rich and deep methane-rich groundwaters induced a sulphate-reducing microbial community using methane and acetate as the key electron and carbon donors in the aquifers of ONK-PVA6.
The construction of the ONKALO tunnel causes a drawdown of sulphate-rich groundwater that mixes with the deep methane-rich groundwater. It has been found that the mixing layer is slowly moving deeper in fractures intersected by the ONKALO tunnel. The ONK-PVA6 borehole was drilled on 3–4 November 2009 and the groundwater at that time was sulphate poor. The sulphate concentration has, due to the drawdown, been slowly rising since then to 1.9 mM, analyzed as of 12 April 2012. The FCCS experiments reported here were designed to mimic this human-induced transition in groundwater geochemistry that is slowly working its way deeper in ONKALO, triggering SRB growth in the mixing layer. Sequences and cultures with sulphate-reducing ability appeared in both groundwater and biofilms in the sulphate-enriched FCCS in similar proportions, i.e., approximately 1% of total numbers of cells and sequences ( and ). Sulphate was consequently the only compound needed to cause SRB growth in ONK-KR15 groundwater. Adding ONK-PVA6 groundwater had only a small effect on the sequence diversity and number of cultivable microorganisms.
The ONK-PVA6 groundwater had sequences representing Desulfobacula, an OTU found in the Sulphate + ONK-PVA6 biofilm but not in the sulphate biofilm. Otherwise, the two treatments did not differ substantially. The effect of sulphate was slow, and it took more than 60 d for the MPN of cultivable SRB to increase (). This coincided with the time when Eh approached –250 mV in the sulphate FCCS, which is a typical Eh at which SRB activity has previously been observed in the FCCSs (Pedersen 2012b). Although no SRB-related sequences were detected in the control biofilm 454 sequence library, there may still have been a few cells of SRB genera in the rare biosphere (Sogin et al. Citation2006) of the FCCSs that could grow after the sulphate treatment. The FCCS experiment accordingly corroborated the hypothesis that sulphate is the only compound needed to invoke SRB growth in sulphate-poor deep groundwater of Olkiluoto.
A high Km for methane of 37 mM has been reported for an anaerobic oxidation of methane (AOM) process in sediments (Zhang et al. Citation2010). If methane was the main electron donor for the SRB community, the observed slow increase in cell numbers and the limited representation of SRB-related OTUs in the sequence libraries of the sulphate-amended FCCSs may relate to such a high Km for AOM with sulphate. Consequently, although AOM with sulphate may have been ongoing in the sulphate-amended FCCSs, the process may have been too slow for detection at the apparent methane concentration of approximately 4.5 mM.
The presence of viruses, i.e., phages that attack microorganisms in groundwater must originate from lytic infections of host microorganisms. Investigation of Äspö Hard Rock Laboratory groundwater for phage abundance returned large numbers of a diverse phage population (Kyle et al. Citation2008). The average ratio of VLP to TNC was 12, indicating active microbial populations. If phages in deep groundwater are active and lytic, they will constitute an important group of predators that might control microorganism numbers and activity. Furthermore, their presence suggests that their prey—the microorganisms—are active and growing. The ratio of VLP to TNC was high during the first half of the experiment, after which it decreased to 2–4 (). This decrease coincided with the increase in NRB and SRB in the sulphate-amended FCCSs ( and ). It seems as the phages initially exerted a significant controlling effect on the numbers of microbial cells, in particular SRB, in the FCCSs.
In conclusion, this work has demonstrated a clear relationship between deep groundwater composition and microbial diversity. The presence/absence of only one geochemical parameter, i.e., sulphate, in the groundwater induced a very large community transition. The investigated subterranean microbial communities consequently had the capacity to respond to changes in the geochemical environment by manifesting community transitions. Both cultivation and 454 pyrotag sequencing indicated SRB communities to be very competitive in the deep biosphere in the presence of sulphate. Because SRB activity generates sulphide that is corrosive to metals, these conclusions should be considered when evaluating the influence of microbial processes on the future geological disposal of radioactive wastes in metal canisters.
Acknowledgments
The pyrotag sequencing data were made possible by the Deep Carbon Observatory's Census of Deep Life program supported by the Alfred P. Sloan Foundation. Pyrotag sequencing was performed at the Marine Biological Laboratory (Woods Hole, MA, USA), where we received excellent assistance from Sharon Grim, Hilary Morrison, Susan Huse, Mitch Sogin, Joseph Vineis, and Andrew Voorhis. The assistance of Maarit Yli-Kaila at the ONKALO site was first rate and indispensable. The authors are grateful to Björn Hallbeck, Jessica Johansson, and Linda Johansson at Microbial Analytics Sweden for their excellent laboratory work.
Funding
The research leading to these results received funding from Posiva Oy.
References
- Aalto P, Helin J, Lindgren S, Pitkänen P, Ylä-Mella M, Ahokas H, Heikkinen E, Klockars J, Lahdenperä A-M, Korkealaakso J, et al. 2011. Baseline Report for Infiltration Experiment. Olkiluoto, Finland: Posiva Oy. Working Report nr WR 2011–25. 152 p.
- Brazelton WJ, Morrill PL, Szponar N, Schrenk MO. 2013. Bacterial communities associated with subsurface geochemical processes in continental serpentinite springs. Appl Environ Microbiol 79:3906–3916.
- Brysch K, Schneider C, Fuchs G, Widdel F. 1987. Lithoautotrophic growth of sulphate-reducing bacteria, and description of Desulfobacterium autotrophicum gen. nov., sp. nov. Arch Microbiol 148:264–274.
- Ekendahl S, Pedersen K. 1994. Carbon transformations by attached bacterial populations in granitic groundwater from deep crystalline bed-rock of the Stripa research mine. Microbiology 140:1565–1573.
- Eydal HSC, Pedersen K. 2007. Use of an ATP assay to determine viable microbial biomass in Fennoscandian Shield groundwater from depths of 3–1000 m. J Microbiol Methods 70:363–373.
- Gihring TM, Moser DP, Lin L-H, Davidson M, Onstott TC, Morgan L, Milleson M, Kieft TL, Trimarco E, Balkwill DL, et al. 2006. The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol J 23:415–430.
- Hallbeck L, Pedersen K. 2008. Characterization of microbial processes in deep aquifers of the Fennoscandian Shield. Appl Geochem 23:1796–1819.
- Hobbie JE, Daley RJ, Jasper S. 1977. Use of nucleopore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol 33:1225–1228.
- Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 14:2317–2319.
- Huse S, Huber J, Morrison H, Sogin M, Welch D. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:R143.
- Jägevall S, Rabe L, Pedersen K. 2011. Abundance and diversity of biofilms in natural and artificial aquifers of the Äspö Hard Rock Laboratory, Sweden. Microb Ecol 61:410–422.
- Kuever J, Könneke M, Galushko A, Drzyzga O. 2001. Reclassification of Desulfobacterium phenolicum as Desulfobacula phenolica comb. nov. and description of strain Sax T as Desulfotignum balticum gen. nov., sp. nov. Int J Syst Evol Microbiol 51:171–177.
- Kyle JE, Eydal HSC, Ferris FG, Pedersen K. 2008. Viruses in granitic groundwater from 69 to 450 m depth of the Äspö hard rock laboratory, Sweden. ISME J 2:571–574.
- Lane DJ. 1991. 16S/23S rDNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester, UK: John Wiley & Sons Ltd. p115–175.
- Lin L-H, Wang P-L, Rumble D, Lippmann-Pipke J, Boice E, Pratt L, Lollar BS, Brodie EL, Hazen TC, Andersen GL, et al. 2006. Long-term sustainability of a high-energy low-diversity crustal biome. Science 314:479–482.
- Marteinsson VT, Rúnarsson A, Stefánsson A, Thorsteinsson T, Jóhannesson T, Magnússon SH, Reynisson E, Einarsson B, Wade N, Morrison HG, et al. 2013. Microbial communities in the subglacial waters of the Vatnajökull ice cap, Iceland. ISME J 7:427–437.
- Motamedi M, Pedersen K. 1998. Desulfovibrio aespoeensis sp. nov., a mesophilic sulfate-reducing bacterium from deep groundwater at Äspö hard rock laboratory, Sweden. Int J Syst Bacteriol 48:311–315.
- Noble RT, Fuhrman JA. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol 14:113–118.
- Pedersen K. 1982. Method for studying microbial biofilms in flowing-water systems. Appl Environ Microbiol 43:6–13.
- Pedersen K. 2005. The MICROBE framework: Site descriptions, instrumentation, and characterization, Äspö Hard Rock Laboratory. Stockholm: Swedish Nuclear Fuel and Waste Management Co. International Progress Report nr IPR-05-05. 85 p.
- Pedersen K. 2012a. Subterranean microbial populations metabolize hydrogen and acetate under in situ conditions in granitic groundwater at 450 m depth in the Äspö Hard Rock Laboratory, Sweden. FEMS Microbiol Ecol 81:217–229.
- Pedersen K. 2012b. Influence of H2 and O2 on sulphate-reducing activity of a subterranean community and the coupled response in redox potential. FEMS Microbiol Ecol 82:653–665.
- Pedersen K. 2013. Metabolic activity of subterranean microbial communities in deep granitic groundwater supplemented with methane and H2. ISME J 7:839–849.
- Pedersen K, Arlinger J, Ekendahl S, Hallbeck L. 1996. 16S rRNA gene diversity of attached and unattached groundwater bacteria along the access tunnel to the Äspö Hard Rock Laboratory, Sweden. FEMS Microbiol Ecol 19:249–262.
- Pedersen K, Arlinger J, Hallbeck A, Hallbeck L, Eriksson S, Johansson J. 2008. Numbers, biomass and cultivable diversity of microbial populations relate to depth and borehole-specific conditions in groundwater from depths of 4 to 450 m in Olkiluoto, Finland. ISME J 2:760–775.
- Pedersen K, Ekendahl S. 1990. Distribution and activity of bacteria in deep granitic groundwaters of southeastern Sweden. Microb Ecol 20:37–52.
- Pedersen K, Ekendahl S. 1992a. Incorporation of CO2 and introduced organic compounds by bacterial populations in groundwater from the deep crystalline bedrock of the Stripa mine. J Gen Microbiol 138:369–376.
- Pedersen K, Ekendahl S. 1992b. Assimilation of CO2 and introduced organic compounds by bacterial communities in groundwater from southeastern Sweden deep crystalline bedrock. Microb Ecol 23:1–14.
- Pedersen K, Hallbeck L, Arlinger J, Erlandson A-C, Jahromi N. 1997. Investigation of the potential for microbial contamination of deep granitic aquifers during drilling using 16S rRNA gene sequencing and culturing methods. J Microbiol Meth 30:179–192.
- Posiva Oy. 2009. Olkiluoto Site Description 2008, Posiva Report nr 2009-1. 725 p. Eurajoki, Finland: Posiva Oy.
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucl Acids Res 35:7188–7196.
- Ravot G, Magot M, Fardeau ML, Patel BK, Thomas P, Garcia JL, Ollivier B. 1999. Fusibacter paucivorans gen. nov., sp. nov., an anaerobic, thiosulfate-reducing bacterium from an oil-producing well. Int J Syst Bacteriol 3:1141–1147.
- Sherwood Lollar B, Frape SK, Weise SM, Fritz P, Macko SA, Welhan JA. 1993. Abiogenic methanogenesis in crystalline rocks. Geochim Cosmochim Acta 57:5087–5097.
- Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103:12115–12120.
- Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JF. 2001. Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67:5750–5760.
- Toropainen V. 2009. Core drilling of drillholes ONK-PVA06 and ONK-PVA07 in ONKALO at Olkiluoto 2009. Working Report nr 2009-121. 61 p. Eurajoki, Finland: Posiva Oy.
- Toropainen V. 2011. Core drilling of drillholes ONK-KR13, ONK-KR154 and ONK-KR15 in ONKALO at Olkiluoto 2010–2011. Working Report nr 2011-39. 127 p. Eurajoki, Finland: Posiva Oy.
- Willems A, Busse J, Goor M, Pot B, Falsen E, Jantzen E, Hoste B, Gillis M, Kersters K, Auling G, et al. 1989. Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydroge-nophaga flava comb. nov. (formerly Pseudomonas flava), Hydrogenophaga palleronii (formerly Pseudomonas palleronii), Hydrogen-ophaga pseudoflava (formerly Pseudomonas pseudoflava and ‘Pseudomonas carboxydoflava’), and Hydrogenophaga taeniospiralis (formerly Pseudomonas taeniospiralis). Int J Syst Bacteriol 39:319–333.
- Zhang Y, Henriet J-P, Bursens J, Boon N. 2010. Stimulation of in vitro anaerobic oxidation of methane rate in a continuous high-pressure bioreactor. Bioresour Technol 101:3132–3138.
