ABSTRACT
Aerobically grown E. coli cells reduced Pd(II) via a novel mechanism using formate as the electron donor. This reduction was monitored in real-time using extended X-ray absorption fine structure. Transmission electron microscopy analysis showed that Pd(0) nanoparticles, confirmed by X-ray diffraction, were precipitated outside the cells. The rate of Pd(II) reduction by E. coli mutants deficient in a range of oxidoreductases was measured, suggesting a molybdoprotein-mediated mechanism, distinct from the hydrogenase-mediated Pd(II) reduction previously described for anaerobically grown E. coli cultures. The potential implications for Pd(II) recovery and bioPd catalyst fabrication are discussed.
Introduction
The microbial reduction of metals and radionuclides has attracted much interest, as it can be potentially harnessed for bioremediation, metal recovery, the fabrication of novel nanobiominerals and even energy generation in biobatteries (Lloyd Citation2003; Lloyd et al. Citation2008; Lovley Citation2006). For example, the sulfate-reducing bacterium (SRB) Desulfovibrio desulfuricans has been shown to use a periplasmic hydrogenase supplied with hydrogen to reduce soluble Pd(II), resulting in the precipitation of Pd(0) nanoparticles in the periplasm of the cell (‘bioPd’). However SRB produce H2S, a potent catalyst poison that must be removed before making the bioPd.
Other organisms capable of this metal bioreduction include the Gram-negative bacteria Shewanella oneidensis (De Windt et al. Citation2005), Escherichia coli (Deplanche et al. Citation2010, Citation2014; Mabbett et al. Citation2006), Pseudomonas putida, Cupriavidus necator (Søbjerg et al. Citation2009), Cupriavidus metallidurans (Gauthier et al. Citation2010), Paracoccus denitrificans (Bunge et al. Citation2010), Rhodobacter sphaeroides (Redwood et al. Citation2008), Rhodobacter capsulatus (Wood et al. Citation2010), and the Gram-positive bacteria Bacillus sphaericus (Creamer et al. Citation2007), Arthrobacter oxyidans (Deplanche et al. Citation2014; Wood et al. Citation2010), Micrococcus luteus (Deplanche et al. Citation2014), Staphylococcus sciuri (Søbjerg et al. Citation2009) and Clostridium pasteurianum (Chidambaram et al. Citation2010).
This property has allowed the use of “palladized” whole cells or processed biomineral directly in industrially important reactions, often showing superior activity compared with a commercially available carbon-supported palladium catalyst. A number of studies have investigated the catalytic activity of bioPd, demonstrating its use in remediative reactions such as the reduction of Cr(VI) to Cr(III) (Beauregard et al. Citation2010; Mabbett et al. Citation2006), the dehalogenation of chlorophenol, polychlorinated biphenyls, polybrominated diphenyl ethers (Baxter-Plant et al. Citation2003; De Windt et al. Citation2005; Harrad et al. Citation2007), trichloroethylene (Hennebel et al. Citation2009a, Citation2009b), and the pesticide γ- hexachlorocyclohexane (Mertens et al. Citation2007), in “greener” chemical synthesis such as the hydrogenation of itaconic acid (Creamer et al. Citation2007) and 2-pentyne (Bennett et al. Citation2010), in Heck and Suzuki reactions (Bennett et al. Citation2013; Deplanche et al. Citation2014), and also in the application of bioPd as a fuel cell electrocatalyst to produce electricity from hydrogen (Orozco et al. Citation2010; Yong et al. Citation2007). In each case where the bioPd was compared with an abiotically produced palladium catalyst (finely divided or supported on a carbon matrix), the bioPd was more active than or at least as active as the commercially available alternative.
Production of catalytically active bioPd also was reported by an aerobically grown Serratia sp. (Beauregard et al. Citation2010; Deplanche et al. Citation2014) under which condition hydrogenases are not expressed. Also, cells of E. coli deficient in the three major hydrogenases reduced Pd(II) (albeit slowly: Deplanche et al. Citation2010), and showed larger Pd-nanoparticles located on the outer surface of the cells. This suggested an alternative mechanism of Pd(II) reduction which has not been investigated.
E. coli produces bioPd which is comparably active to that produced by D. desulfuricans (Deplanche et al. Citation2014). This also provides a very useful model organism since it is facultatively anaerobic and has well-defined molecular tools to elucidate reaction mechanisms under aerobic and anaerobic conditions. The enzymes potentially involved in the bioreduction of palladium by E. coli under the latter conditions are the nickel-dependent hydrogenase enzymes Hyd-1, Hyd-2, and Hyd-3, and the formate dehydrogenase molybdoenzymes FDH-N, and FDH-H. Another molybdoenzyme, FDH-O, is expressed under both aerobic and anaerobic conditions.
A possible role for FDH-O is to allow bacteria to adapt rapidly to a sudden shift from aerobic respiration to anaerobiosis, before FDH-N has been produced in sufficient amounts to continue formate metabolism (Abaibou et al. Citation1995). Hyd-1, Hyd-2, FDH-O, and FDH-N are membrane-bound and periplasmically oriented, whereas Hyd-3 and FDH-H are subunits of the formate hydrogenlyase (FHL) complex, an intracellular enzyme complex that is also membrane-bound but which faces into the cytoplasm. The mechanisms responsible for the formate-dependent bioreduction by anaerobically grown cultures of E. coli have been studied, showing that the hydrogenase enzymes Hyd-1 and Hyd-2 are mainly responsible for Pd(II) bioreduction (Deplanche et al. Citation2010). In a study of formate-dependent Pd(II) bioreduction by Desulfovibrio fructosovorans, the deletion of the periplasmic hydrogenases caused the Pd(0) nanoparticles to be relocated to the cytoplasmic membrane site of the remaining hydrogenases, indicating that the periplasmic hydrogenases are at least partially involved (Mikheenko et al. Citation2008).
The growth yield of anaerobic cultures is lower than that of aerobic cultures, and for economic production at scale a method of growth of high biomass density is required. When using anaerobic cultures there is also the cost of supplementing with sodium fumarate and glycerol. The dual aims of this study are to establish whether E. coli cells grown aerobically are capable of manufacturing bioPd and to identify the enzyme(s) responsible for such metal reduction. A move away from the need for anaerobic growth would simplify the preparation of high levels of active biomass for catalyst production at industrial scale.
Methods
Bacterial growth
Starter cultures: 50 ml LB broth in a 500-ml Erlenmeyer flask was inoculated with a single isolated colony of the E. coli strain under investigation and incubated aerobically (37°C, shaking at 180 rpm for 18 h).
Aerobic cultures
An 11-ml starter culture was added to 99 ml LB broth in a 1-L Erlenmeyer flask. Flasks were incubated for 24 h (37°C, 180 rpm) to produce stationary phase ‘resting’ cells. The pH of the cells after 24-h incubation was measured to determine that organic acids had not been produced that would otherwise lower the pH considerably (Vasala et al. Citation2006). Oxygen saturation of a 5-ml aliquot of the broth culture was measured immediately after 24 h of incubation using an Oakton D06 Acorn Series dissolved oxygen meter.
Reduction of Pd(II) to produce bioPd on bacteria
The aerobically grown liquid culture was divided between two 50-ml Falcon tubes and washed three times in 20 ml MOPS-NaOH (morpholinepropanesulfonic acid) buffer, 20 mM at pH7.6 after centrifugation for 20 min at 2500 g. Cell pellets were adjusted to a mass of 250 mg wet pellet weight, and resuspended in the MOPS-NaOH buffer to a volume of 1 ml. One tube of 250 mg wet weight cells was resuspended in 22.5 ml MOPS-NaOH buffer with 1 mM sodium tetrachloropalladate in a 30 ml bottle sealed with a butyl rubber stopper. The bottle was incubated in the dark at 30°C for 1 h for the Pd(II) to biosorb to the cells (Baxter-Plant et al. Citation2003). Then 2.5 ml 10 mM sodium formate was then added to the bottle to initiate bioreduction of the Pd(II).
Use of mutants to determine electron transfer pathway to Pd(II)
In order to investigate the possible role of the aerobic formate dehydrogenase (FDH-O) and other hydrogenase/formate dehydrogenase enzymes in the reduction of Pd(II) by aerobically grown cells of E. coli, the rates of reduction by six different additional strains () were compared by measuring the Pd(II) remaining in solution by ICP-MS. The strains were “palladized” as above, and rates of reduction/removal compared to those in a series of controls: killed cells (MC4100), cell-free suspension, and live cells (MC4100) unsupplemented with formate.
Table 1. E. coli strains used to determine biological involvement in the reduction of palladium (II) using formate as the electron donor.
All strains except BL21(DE3) were from the culture collection of Professor Frank Sargent at the College of Life Sciences, University of Dundee. Strain BL21(DE3) was obtained from Invitrogen, Paisley, UK. Strain MC4100 ΔmoaA was created by disruption of the moaA gene, which encodes the molybdenum cofactor biosynthesis protein A, using the method of Datsenko and Wanner (Citation2000) whereby PCR products are used to disrupt the gene of choice by recombination using the plasmid-borne phage λ Red recombinase.
X-ray diffraction (XRD) analysis
The black precipitates were washed once in acetone and air dried before analysis by X-ray diffraction (XRD). The measurements were performed on a Bruker D8 Advance diffractometer, using Cu κ-α1 radiation. The samples were scanned from 5–70 degrees 2θ in steps of 0.2 degrees, with a count time of 2 sec per step.
Extended X-ray absorption fine structure (EXAFS)
Aliquots of the cell/Pd/formate suspension were taken at times 0 and 30 min, and 1, 3 and 4 h from the addition of formate, and frozen immediately in liquid nitrogen. The direct reduction of Pd(II) to Pd(0) was demonstrated using EXAFS, performed at the European Synchrotron Radiation Facility (ESRF), in Grenoble, France. The samples were transported to the synchrotron at ESRF on dry ice, where they were thawed and injected immediately into sample holders, before freezing once more in liquid nitrogen and placing into the beam. X-ray absorption data were collected on beamline BM29 at the Pd K–edge in the energy range 24,200–24,900 eV.
Data were recorded at low temperature (77 K) and under vacuum to reduce the thermal Debye-Waller factor and prevent oxidation. A Si(III) double crystal monochromator was used, calibrated with a Pd foil, and the spectra were collected in fluorescence mode using a 13-element solid-state detector. A reference spectrum of a palladium foil was recorded in transmission mode on station 9.3 at the SRS Daresbury. The data were background subtracted and the EXAFS spectra fitted in DL Excurv (http://www.cse.scitech.ac.uk/cmg/EXCURV/) using full curved wave theory (Gurman et al. Citation1984).
Transmission electron microscopy (TEM) and energy dispersive X-ray spectroscopy (EDS)
Following Pd(II) reduction, cells were stored at 10°C overnight. The cell pellets were then rinsed twice with deionized water, fixed in 2.5% (wt/vol) glutaraldehyde, centrifuged for 5 min at 16,000 g, resuspended in 1.5 ml of 0.1 M cacodylate buffer (pH 7) and stained in 1% osmium tetroxide in 0.1 M phosphate buffer, pH 7 (60 min). Cells were dehydrated using an ethanol series (70, 90, 100, 100, 100% dried ethanol, 15 min each) and washed twice in propylene oxide (15 min, 9500 g). Cells were embedded in epoxy resin and the mixture was left to polymerize (24 h; 60°C). Sections (100–150-nm thick) were cut from the resin block, placed onto a copper grid and viewed with a JEOL 1200CX2 TEM, accelerating voltage 80 keV. EDS was performed on electron-dark areas, to confirm the presence of palladium.
Results
Palladization of E. coli BL21(DE3)
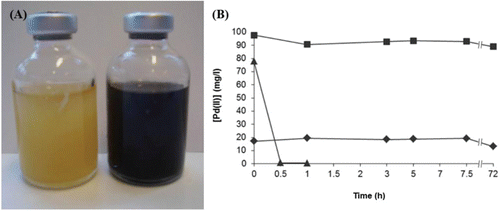
The pH of the aerobically grown liquid culture was between 7.7–7.9, indicating that there was not extensive production of organic acids due to overflow metabolism. Oxygen saturation measurements showed that the liquid culture was 72% saturated following 24 h of incubation, indicating that it was not oxygen-limited. After harvesting, the cells were able to couple the reduction of Pd(II) to the oxidation of formate, indicated by the rapid formation of a black precipitate, tentatively identified as Pd(0) (). ICP-MS analysis confirmed complete removal of Pd(II) from solution within 45 min, and the presence of crystalline Pd(0) was confirmed using XRD in this, but not in the heat-killed cells control where the cells removed substantial Pd(II) abiotically. An increase in metal biosorption by heat killed biomass as compared to live cells is well documented (Machado et al. Citation2009; Parameswari et al. Citation2009) and was attributed to loss of membrane integrity to reveal additional intracellular metal binding sites (Machado et al. Citation2009).
Figure 1. (A) Complete reduction of Pd(II) to Pd(0) by an aerobically grown culture of E. coli. Both bottles contain cells resuspended in 20 mM MOPS buffer at pH 7.6, and 1 mM sodium tetrachloropalladate (total volume 25 ml). This image was taken 45 min after the addition of formate to the bottle on the right. (B) Reduction by E. coli MC4100 and by controls showing no abiotic reduction of Pd(II). Controls used were killed (autoclaved) cells and cell-free suspension. Soluble Pd(II) in the supernatant was measured using ICP-MS. ▴ = MC4100; ■ = no cells; ♦ = killed cells.
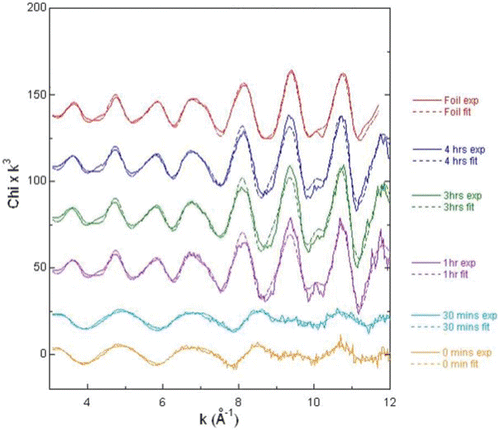
Extended X-ray absorption fine structure (EXAFS)
The nature of the Pd associated with the biomass was assessed further using X-ray absorbance spectroscopy. The features in the corresponding EXAFS spectra () are due to the wavelike nature of the photoelectron, which is released from the atom with increasing energy and scattered from surrounding atoms with new waves being emitted. With increasing photon energy, the interference between the waves alternates between constructive and destructive, which leads to oscillations in the spectrum. Examining these oscillations gives information on the number, species and distance of the surrounding atoms. As seen in , the samples taken at times 0 and 30 min, which contain Pd(II), have identical EXAFS spectra. The samples taken at 60 min onwards are identical to the Pd(0) foil control, which indicates that only Pd(0) was present. Reduction of the Pd(II) to Pd(0) was therefore confirmed to be complete in less than 30 min, as confirmed by ICP-MS analysis.
Use of mutants to determine electron transfer pathway to Pd(II)
Aerobic cultures of the parental strains MC4100 and BW25113 and the strain which lacked all hydrogenases (JW2682) removed Pd(II) identically with no residual Pd(II) detected after 30 min (). Removal of the hydrogenase enzymes had no effect on the rate of palladium removal from solution, confirming that these hydrogenases have no role in the aerobic reduction of Pd(II). The FDH-O-negative strain JW3865 reduced Pd(II) within 1 h, and the FDH-O/FDH-N-negative strain FTD128 within 2 h. Strain MC4100 ΔmoaA, lacking all molybdoenzymes, reduced the palladium within 7 h.
Figure 3. Pd(II) reduction by six different strains of E. coli, using formate as the electron donor. Soluble Pd(II) in the supernatant was measured using ICP-MS. ♦ = BW25113; □ = JW2682; ▴ = JW3865; Δ = MC4100 ΔmoaA; ■ = MC4100; ◊ = FTD128. Data points for BW25113, JW2682 and JW3865 are mean values of triplicates, with standard error shown.
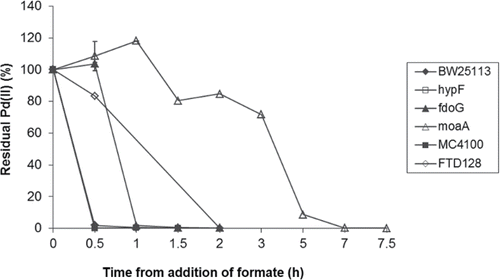
These results indicate the likely involvement of the FDH-O enzyme in the reduction of Pd(II) by aerobically grown E. coli using formate, although other Mo-containing enzymes must also be involved given the impaired metal reduction noted with the Δmoa mutant. Controls containing no biomass showed no abiotic reduction of Pd(II) using formate (), although a brown precipitate was seen in the no-formate control. The X-ray powder diffraction pattern did not show the presence of any peaks characteristic of Pd(0) in this precipitate, indicating that it was probably amorphous and noncrystalline. Time zero on is the point at which formate was added, following 1 h of incubation to allow biosorption of the Pd(II) to the cells; hence the abiotic Pd(II) removal by killed cells () was apparent at the time of formate addition with no evidence for further Pd(II) reduction.
Transmission electron microscopy (TEM)
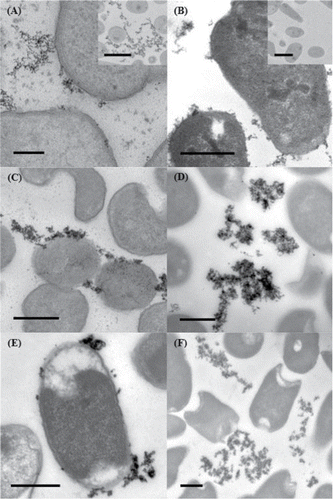
TEM images of thin sections of cells showed that with all strains the reduced palladium was precipitated predominantly in the extracellular matrix of the cultures (), although it appears that the nanoparticles may be associated with the outer membrane of the cells. Energy dispersive X-ray spectroscopy (EDS) confirmed the presence of palladium in these precipitates.
Discussion
The results from this study demonstrate that it is possible for aerobically grown cultures of E. coli to reduce Pd(II) enzymatically, with no need to remove oxygen from the experimental system during the bioreduction step. Autoclaved control experiments indicate that Pd(II) bioreduction in these cultures is enzymatic, with reduction of palladium not occurring in the absence of viable cells irrespective of the length of incubation. The major enzymes shown to be involved include the formate dehydrogenases FDH-O and FDH-N, although bioreduction still occurs in strains without these enzymes albeit at a much lower rate. Other molybdoenzymes must therefore be involved.
The strain that lacked all molybdoenzymes did however still reduce the palladium, although this took 7 h, compared with less than 30 min by the wild-type strains. Hydrogenases, implicated as the dominant Pd(II) reductases in other experimental systems grown under anaerobic conditions (Deplanche et al. Citation2010; Mikheenko et al. Citation2008), are not expressed in aerobically grown cultures, and their lack of involvement was evident as the strain lacking hydrogenase enzymes reduced palladium at the same rate as the wild-type strains in this study.
Furthermore, whichever biological system is responsible for the aerobic bioreduction of Pd(II), there seems to be little impact on the site of Pd(0) deposition. The location of the bioreduced Pd(0) in our experiments is almost always extracellular, although often associated with the outer membrane of the cells. This is particularly the case with the MC4100 ΔmoaA strain (which lacks all molybdoenzymes), in which the majority of the Pd(0) nanoparticles are closely associated with the outer membrane ().
One conclusion that may be drawn from this is that whilst cells that lack the formate dehydrogenases are still capable of reducing Pd(II), when all of these enzymes are missing a cellular component associated with the outer membrane may be responsible. Furthermore, this formate oxidation activity is much weaker than that seen with the strains containing formate dehydrogenases, where Pd(II) reduction is more rapid. It is possible however that following the initial enzymatic reduction of a small percentage of the Pd(II), the Pd(0) nanoparticles formed may themselves be responsible for catalysing the reduction of the remainder of the Pd(II) (Yong et al. Citation2002), which would mean that only a minor, initial biological input is required.
Although the formate dehydrogenase enzyme systems implicated in Pd(II) bioreduction by E. coli are periplasmic, the majority of the reduced Pd(0) precipitates outside the cell. It is possible that an electron shuttle system exists similar to that found in Shewanella oneidensis (von Canstein et al. Citation2008) that is as yet undiscovered in E. coli. It is also possible that the first Pd(0) nanoparticles to form breach the outer membrane, and themselves form an electron conduit for further Pd(II) reduction outside the cell. The pH of these experiments is also higher than others where Pd(0) nanoparticles accumulated in the periplasm (Redwood et al. Citation2008), which could indicate the higher biosorption of cationic metal to the outer membrane and extracellular polymeric substances, which are then not able to enter the periplasm. The influence of a higher pH in the location of the Pd(0) may be confirmed by the observation that Pd(0) nanoparticles were located on the cell surface of D. desulfuricans when bioreduction of Pd(II) was performed at pH 7 (Yong et al. Citation2002).
In conclusion, this study has demonstrated the presence of a novel biological mechanism responsible for the bioreduction of Pd(II) in aerobically grown cultures of E. coli, catalyzed mainly by molybdenum-containing enzyme systems. Subsequent studies will investigate the catalytic activity and selectivity of the Pd(0) nanoparticles produced under aerobic conditions in a range of industrially important reactions. If active, this new form of bioPd has the advantage over that produced by anaerobic culture as it is easier to produce at high yield, from increased biomass levels associated with aerobic growth.
There is also no requirement for additional processing steps to remove H2S (produced by SRB systems), and the use of formate instead of hydrogen gas means that the procedure is less hazardous and more controllable. The advantages of this more scalable method of synthesis would need to be considered against any alterations in activity/selectivity of the resulting catalyst (versus synthetic and other bioPds), using a cost-benefit analysis. Importantly, identification of the specific enzymatic process(es) involved in the biomanufacture of bioPd is the first step towards application of the tools of synthetic biology for “designer catalyst” production for specific applications.
In a geomicrobiological context, this study shows that aerobic cells of E. coli restrict the deposition of Pd(0) to locations outside the cell. However in both D. desulfuricans (grown anaerobically) and Bacillus benzeovorans (grown aerobically) intracellular depositions of small Pd-nanoparticles were observed at the expense of both hydrogen and formate (Omajali, Mikheenko, Merroun, Wood, and Macaskie in press) and, notably, were also seen in E. coli grown anaerobically (Macaskie, Williams, Priestley, and Courtney unpublished). This raises questions about potential biochemical ‘trafficking’ pathways of Pd(II), the possibility of Pd(II) efflux by aerobic (but not anaerobic) cells and, following from that, the possibility of biogeochemical cycling of this element.
Acknowledgment
The authors would like to thank Dr Victoria Coker, Prof Richard Pattrick, and Dr John Charnock of the University of Manchester for assistance with EXAFS.
Funding
This study was supported by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC).
References
- Abaibou H, Pommier J, Benoit S, Giordano G, Mandrandberthelot MA. 1995. Expression and characterization of the Escherichia coli fdo Locus and a possible physiological role for aerobic formate dehydrogenase. J Bacteriol 177:7141–7149.
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2: Article 2006.0008.
- Baxter-Plant V, Mikheenko IP, Macaskie LE. 2003. Sulphate-reducing bacteria, palladium and the reductive dehalogenation of chlorinated aromatic compounds. Biodegradation 14:83–90.
- Beauregard D, Yong P, Macaskie LE, Johns ML. 2010. Using non-invasive magnetic resonance imaging (MRI) to assess the reduction of Cr(VI) using a biofilm-palladium catalyst. Biotechnol Bioeng 107:11–20.
- Bennett JA, Creamer NJ, Deplanche K, Macaskie LE, Shannon IJ, Wood J. 2010. Palladium supported on bacterial biomass as a novel heterogeneous catalyst: A comparison of Pd/Al2O3 and bio-Pd in the hydrogenation of 2-pentyne. Chem Eng Sci 65:282–290.
- Bennett JA, Mikheenko I, Deplanche K, Shannon IJ, Wood J, Macaskie LE. 2013. Nanoparticles of palladium supported on bacterial biomass: new re-usable heterogeneous catalyst with comparable catalytic activity to homogeneous colloidal palladium in the Heck reaction. Appl Catal B Environ 140–141:700–707.
- Bunge M, Søbjerg LS, Rotaru AE, Gauthier D, Lindhardt AT, Hause G, Finster K, Kingshott P, Skrydstrup T, Meyer RL. 2010. Formation of palladium (0) nanoparticles at microbial surfaces. Biotechnol Bioeng 107:206–215.
- Casadaban MJ, Cohen SN. 1979. Lactose genes fused to exogenous promoters in one-step using a Mu-lac Bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA 76:4530–4533.
- Chidambaram D, Hennebel T, Taghavi S, Mast J, Boon N, Verstraete W, van der Lelie D, Fitts JP. 2010. Concomitant microbial generation of palladium nanoparticles and hydrogen to immobilize chromate. Environ Sci Technol 44:7635–7640.
- Creamer NJ, Mikheenko IP, Yong P, Deplanche K, Sanyahumbia D, Wood J, Pollmann K, Merroun M, Selenska-Pobell S, Macaskie LE. 2007. Novel supported Pd hydrogenation bionanocatalyst for hybrid homogeneous/heterogeneous catalysis. Catal Today 128:80–87.
- Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645.
- De Windt W, Aelterman P, Verstraete, W. 2005. Bioreductive deposition of palladium (0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls. Environ Microbiol 7:314–325.
- Deplanche K, Bennett JA, Mikheenko IP, Omajali J, Wells AS, Meadows PE, Wood J, Macaskie LE. 2014. Catalytic activity of biomass supported palladium nanoparticles: influence of the biological component in catalytic efficacy and potential applications in green synthesis of fine chemicals and pharmaceuticals. Appl Catal B Environ 147:651–665.
- Deplanche K, Caldelari I, Mikheenko IP, Sargent F, Macaskie LE. 2010. Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains. Microbiology 156:2630–2640.
- Gauthier D, Søbjerg LS, Jensen KM, Lindhardt AT, Bunge M, Finster K, Skrydstrup T, Meyer RL. 2010. Environmentally benign recovery and reactivation of palladium from industrial waste by using Gram-negative bacteria. Chemsuschem 3:1036–1039.
- Gurman SJ, Binsted N, Ross I. 1984. A rapid, exact curved-wave theory for EXAFS calculations. J Phys C: Solid State Phys 17:143–151.
- Harrad S, Robson M, Hazrati S, Baxter-Plant VS, Deplanche K, Redwood MD, Macaskie LE. 2007. Dehalogenation of polychlorinated biphenyls and polybrominated diphenyl ethers using a hybrid bioinorganic catalyst. J Environ Monitor 9:314–318.
- Hennebel T, Simoen H, De Windt W, Verloo M, Boon N, Verstraete W. 2009b. Biocatalytic dechlorination of trichloroethylene with bio-palladium in a pilot-scale membrane reactor. Biotechnol Bioeng 102:995–1002.
- Hennebel T, Verhagen P, Simoen H, De Gusseme B, Vlaeminck SE, Boon N, Verstraete W. 2009a. Remediation of trichloroethylene by bio-precipitated and encapsulated palladium nanoparticles in a fixed bed reactor. Chemosphere 76:1221–1225.
- Lloyd JR. 2003. Microbial reduction of metals and radionuclides. FEMS Microbiol Rev 27:411–425.
- Lloyd JR, Pearce CI, Coker VS, Pattrick RA, van der Laan G, Cutting R, Vaughan DJ, Paterson-Beedle M, Mikheenko IP, Yong P, Macaskie LE. 2008. Biomineralization: linking the fossil record to the production of high value functional materials. Geobiology 6:285–297.
- Lovley DR. 2006. Microbial fuel cells: novel microbial physiologies and engineering approaches. Curr Opin Biotechnol 17:327–332.
- Luke I, Butland G, Moore K, Buchanan G, Lyall V, Fairhurst SA, Greenblatt JF, Emili A, Palmer T, Sargent F. 2008. Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch Microbiol 190:685–696.
- Mabbett A, Sanyahumbi D, Yong P, Macaskie LE. 2006. Biorecovered precious metals from industrial wastes: Single-step conversion of a mixed metal liquid waste to a bioinorganic catalyst with environmental application. Environ Sci Technol 40:1015–1021.
- Machado MD, Janssens S, Soares HMVM, Soares EV. 2009. Removal of heavy metals using a brewers' yeast strain of Saccharomyces cerevisiae: advantages of using dead biomass. J Appl Microbiol 106:1792–1804.
- Mertens B, Blothe C, Windey K, De Windt W, Verstraete W. 2007. Biocatalytic dechlorination of lindane by nano-scale particles of Pd(0) deposited on Shewanella oneidensis. Chemosphere 66:99–105.
- Mikheenko IP, Rousset M, Dementin S, Macaskie LE. 2008. Bioaccumulation of palladium by Desulfovibrio fructosivorans wild-type and hydrogenase-deficient strains. Appl Environ Microbiol 74:6144–6146.
- Orozco RL, Redwood MD, Yong P, Macaskie LE. 2010. Towards an integrated system for bio-energy: hydrogen production by Escherichia coli and use of palladium-coated waste cells for electricity generation in a fuel cell. Biotechnol Lett 32:1837–1845.
- Paramewsari E, Lakshmanan A, Thilagavathi T. 2009. Effect of pretreatment of blue green algal biomass on biosorption of chromium and nickel. J Algal Biomass Util 1:9–17.
- Redwood MD, Deplanche K, Baxter-Plant VS, Macaskie LE. 2008. Biomass-supported palladium catalysts on Desulfovibrio desulfuricans and Rhodobacter sphaeroides. Biotechnol Bioeng 99:1045–1054.
- Søbjerg LS, Gauthier D, Lindhardt AT, Bunge M, Finster K, Skrydstrup T, Meyer RL. 2009. Bio-supported palladium nanoparticles as a catalyst for Suzuki-Miyaura and Mizoroki-Heck reactions. Green Chem 11:2041–2046.
- Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol 189:113–130.
- Vasala A, Panula J, Bollok M, Illmann L, Halsig C, Neubauer P. 2006. A new wireless system for decentralized measurement of physiological parameters from shake flasks. Microb Cell Factor 5:8.
- von Canstein H, Ogawa J, Shimizu S, Lloyd JR. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl Environ Microbiol 74:615–623.
- Wood J, Bodenes L, Bennett J, Deplanche K, Macaskie LE. 2010. Hydrogenation of 2-butyne-1,4-diol using novel bio-palladium catalysts. Indust Eng Chem Res 49:980–988.
- Yong P, Paterson-Beedle M, Mikheenko IP, Macaskie LE. 2007. From bio-mineralization to fuel cells: biomanufacture of Pt and Pd nanocrystals for fuel cell electrode catalyst. Biotechnol Lett 29:539–544.
- Yong P, Rowson N, Farr JPG, Harris IR, Macaskie, LE. 2002. Bioreduction and biocrystallization of palladium by Desulfovibrio desulfuricans NCIMB 8307. Biotechnol Bioeng 80:369–379.