Abstract
Since the late 1970s, allozymes, microsatellites, and DNA sequence variation have been widely used to study the taxonomy, ecology, and evolution of mayflies (Ephemeroptera). Early research investigated species limits in morphologically cryptic groups and attempted to link immature and adult stages using genetic markers. Population genetic studies soon followed, with a strong emphasis on measuring gene flow as a proxy for dispersal. Most recently, direct sequencing of DNA and molecular phylogenetic tools have broadened our understanding of mayfly systematics and evolution. Interestingly, these newer approaches have also begun to re-address some of the earliest questions about species circumscription and taxonomy. Here we present an overview of genetics in mayfly research to date, focusing on how genetic approaches have been applied to taxonomy, phylogenetics, and population genetics. We identify a number of outstanding questions and highlight some of the most interesting topics for future research. These include the need for evolutionarily valid species concepts when applying genetics to taxonomy, a better understanding of how parthenogenesis may affect population genetic structure, and increased use of DNA sequence data from nuclear gene regions.
Introduction
Laboratory techniques for measuring genetic polymorphism at the biochemical or molecular level have provided significant advances to our understanding of organismal ecology and evolution. Allozyme electrophoresis was the first widely adapted tool for measuring protein polymorphism, and genetic techniques now include the routine identification of polymorphism at individual nucleotides. These may be located anonymously throughout the genome (e.g., RAPD, AFLP, SNP), in microsatellite repeats, or within targeted regions of ribosomal and protein-coding DNA (RFLP, DNA sequencing). Since the earliest study of which we are aware, that of allozyme polymorphism in Baetis harrisoni by Agnew (Citation1978), nearly all of these techniques have been used to study the ecology and evolution of mayflies (Ephemeroptera).
In some of the earliest reports, Saura et al. (Citation1979) found Leptophlebia marginata that swarmed in trees to have different allele frequencies at two allozyme loci compared to those that swarmed above stones. Kownacki and Starmach (1984) found differing patterns of variation in esterase proteins among eight morphological species of mayfly. They suggested this variation could be useful for identification. Zurwerra et al. (Citation1984) used variation at 17 protein loci to distinguish four species of Epeorus. The results of these early studies provide, in hindsight, an accurate prediction of the state of mayfly genetics now, some 24 years later. For example, species that appear to be closely related based on morphological characters had extensive genetic differences at neutral loci (in this case Baetis alpinus and B. melanonyx, and the two Baetidae surveyed were markedly different from the other mayflies (Kownacki and Starmach 1984). We provide examples of how these early conclusions have been repeatedly supported and increasingly refined by more recent phylogenetics research. A third observation based on these earlier works is that no explicit criteria were employed to establish species status using genetic data. We discuss how this shortcoming remains problematic in the more recent literature, and suggest appropriate criteria that could be employed in future genetic work on mayflies.
In the following sections, we attempt to review the body of work that has applied genetic tools to the study of taxonomy, phylogenetics, and ecology of mayflies. Our goal is to review the major findings, highlight some of the shortcomings of genetics tools, and to suggest future directions in mayfly research. Our review is not comprehensive and is admittedly subjective. Faced with limits of time and space, we could not include all references, but our intention is not to minimise other work. Each of the topics we review has been studied with non-genetic methods. For example, there were large bodies of literature on dispersal and systematics before genetic techniques were ever applied to these fields. Here we only mention a few such studies for comparison. In earlier drafts we used expressions like “the first study” or “the earliest work”, but more than once we found an older reference, thus we make no claim to have uncovered the earliest examples of anything. Finally, the methods themselves are not presented in detail; for further information we recommend standard works (e.g., Hartl and Clark Citation1997; Page and Holmes Citation1998; Felsenstein Citation2004).
Taxonomy and species delineation
Genetic distances and fixed character differences
The majority of early applications of genetic techniques to mayfly biology were used in the hope of resolving taxonomic difficulties (Kownacki and Starmach 1984; Zurwerra et al. Citation1984). These included testing hypotheses of species membership (i.e., whether a group of studied specimens constitutes one or more species) and assigning unknown individuals to named species. In most cases, allozyme variation was used to distinguish species that were not easily separated by morphological features. Zurwerra et al. (Citation1984) measured variation at 17 protein loci in four species of Epeorus. Using allele frequency data to generate similarity-based dendrograms, they found all members of a species had a genotype identity (I) approximately equal to 1, meaning that all individuals had identical or nearly identical genotypes at all loci examined. For the different species within a genus, I = 0.33, i.e. species differed by approximately 30%. Finally, they called for more data to corroborate their findings.
Other studies have tried to test for the presence of multiple species within a single Linean binomial using allozymes. Saura et al. (Citation1979) suggested that behaviourally different groups of Leptophlebia marginata had differences in allele frequencies at two allozyme loci. Sweeney and Funk (Citation1991) used allozymes to argue that there were two species within Dolania americana. They cited a similarity of I = 0.95 within these species and also found significant differences in allele frequencies at two allozyme loci. Following from these and other studies, a mixture of allelic differences and genetic similarity have both been used repeatedly to justify species status. Examples include Rhithrogena (Heptageniidae) endemic to Corsica (Belfiore et al. Citation1992), Electrogena (Heptageniidae) from Central Italy (Belfiore et al. Citation1997), and Centroptilum (Baetidae) from eastern North America (Funk et al. Citation2006). The approach has also been used for the generic level. Electrogena (Zurwerra and Tomka Citation1985) was established within the difficult “lateralis group” of the European Heptageniidae, based on 0.2 < I <0.3 indicating the generic rank of all “lateralis group” members.
While many workers have found a high empirical correlation between genetic divergence and taxonomic level (Thorpe Citation1982), the use of distanced-based criteria in taxonomy is problematic for several reasons. First, taxonomy typically classifies organisms according to diagnostic characters, thus using the magnitude of difference in a given set of characters as a criterion is inconsistent (Vogler and Monaghan Citation2007). Diagnostic approaches to species circumscription are well developed for genetic data (Sites and Marshall Citation2004) and have been successfully employed in mayflies. Zloty et al. (Citation1993) used fixed differences at three allozyme loci to distinguish among seven species of Ameletus. They did not present or invoke distance criteria. Funk et al. (Citation2008) found fixed allelic differences among co-occurring morphotypes of Drunella lata, with subsequent analysis uncovering fixed morphological differences as well as ecological differences. A number of the studies cited above also report fixed allelic differences along with genetic distances.
A second concern of distance-based criteria is that there is no basis in evolutionary theory for all species to be similarly different, or, for genetic markers incorporating fixed differences at neutral loci, to be of similar age to one another. Nonetheless, this is what is implied when a similar magnitude of divergence within and among species is invoked as a species criterion. While selective genetic sweeps may normalise the depth of subdivision at some genetic loci (Bazin et al. Citation2006), it is not surprising that the approach yields mixed results (Vogler and Monaghan Citation2007). Another considerable problem is that distance criteria and calibration have the potential to be circular. If a priori species designations are inaccurate, using these to ‘calibrate’ typical thresholds is likely to lead to a confused picture. This may be especially problematic for poorly known groups where very few species have been circumscribed. Finally, genetic distance is a summary statistic that is dependent on the number of loci examined and their level of polymorphism. This was noted in the studies of Saura et al. (Citation1979) and Sweeney and Funk (Citation1991).
Allozymes also potentially suffer from the fact that loci may be expressed differently depending on the environment or developmental stage. Esterase mobility did not change throughout the year in Baetis alpinus and Rhithrogena loyolaea (Kownacki and Starmach 1984), between cohorts of Dolania spp (Funk and Sweeney Citation1991), and between adults and larvae of four species (Zloty et al. Citation1993). In contrast, Scillitani et al. (Citation1996) found variable patterns of allozyme expression in different life stages of Ecdyonurus and Electrogena. The latter finding is difficult to explain but selection of particular allozyme loci has been documented in natural populations of mayflies (Snyder and Hendricks Citation1997). Selection could potentially lead to misdiagnosis of cryptic species if loci are expressed locally.
DNA sequences
Direct sequencing of one or more gene loci has a number of advantages over allozymes for taxonomy and species circumscription. Sequencing of standard regions of DNA, such as cox1 (e.g., Ball et al. Citation2005), reduces the problems of assaying expressed loci and also reduces the variation in descriptive statistics by employing a standardised locus. A more important advantage is that using standardised data in each new study will facilitate a comparative analysis of all previous data. Any new gene sequences can be aligned and compared with any previously published data, meaning that taxonomists will be able to compare their new taxon to existing species and classifications. This will reduce the number of ‘new’ species to be synonymised by future taxonomists (Godfray Citation2002). An unfortunate by-product may be the temptation to interpret cox1 neighbour-joining trees as phylogenetic species trees, against which we strongly caution.
Landolt (Citation1991) used RFLP to assay DNA sequence polymorphism within and among taxonomically difficult Ecdyonurus, and many workers have since employed direct sequencing. While DNA sequences have the potential to eliminate some of the above challenges, any reader who has followed the ‘DNA barcoding’ and ‘DNA taxonomy’ literature will be aware that the most important problems remain (reviewed by Vogler and Monaghan Citation2007). Like the earlier allozyme studies, DNA-based approaches tend to observe distinct visual clusters on a phylogenetic tree or dendrogram. And like allozyme studies, interpretation of these groups remains subject to circularity and to the absence of an evolutionary theory for why genetic distances might be equally different between all species in a given lineage. Williams et al. (Citation2006) observed distinct groups of mtDNA sequences in Baetis rhodani on a phylogenetic tree, but concluded that the species status was unsure. This was arguably the only appropriate conclusion in the absence of a clear and reproducible species concept. In the absence of such a concept, the approach can only be used as a proxy for species boundaries which should then be tested using other criteria, such as ecology (lotic and lentic Baetis macani; Savolainen et al. Citation2007) or morphology (Dipteromimus in Tojo and Matsukawa Citation2003; Centroptilum in Funk et al. Citation2006; Heptagenia in Webb et al. Citation2007).
Our intention is not to be critical of any of the above studies, but to establish the fact that newer genetic methods do not necessarily lead to clearer results in the absence of an objective and repeatable species concept. This is a problem shared by all taxonomic groups and is in no way restricted to the mayflies. Distance-based criteria for species have been problematic in microbial taxonomy for a long time, with a great deal of ecological diversity often found within single distance-based taxa (e.g. Forney et al. Citation2004). Well-established quantitative methods for species delineation exist (e.g. Sites and Marshall Citation2004), although they draw inferences based on a priori groups whose existence is subsequently tested (Davis and Nixon Citation1992). These groups are not always straightforward to establish, particularly in the cases of sympatric, morphologically cryptic species. We argue below (Future perspectives) that this problem can be overcome with existing techniques that employ more advanced analyses and that the use of distance criteria as a species proxy also greatly underutilises the potential of DNA sequences for species circumscription and identification.
Phylogenetics
Phylogeny of the mayflies
The contribution of genetics data to the study of evolutionary relationships within the mayflies has increased greatly since 2000. First, repeated observations from studies of the basal relationships among insect orders are that the Ephemeroptera are monophyletic and that the Baetidae are the sister taxon to all other mayflies (Hovmöller et al. Citation2002; Ogden and Whiting Citation2003). It is not trivial that molecular characters recover the mayflies as monophyletic, considering the inherent difficulties of reconstructing such old lineages with limited data (e.g., Whitfield and Kjer Citation2008). This repeated finding lends strong support to the ability of molecular phylogenetics to reconstruct mayfly evolution.
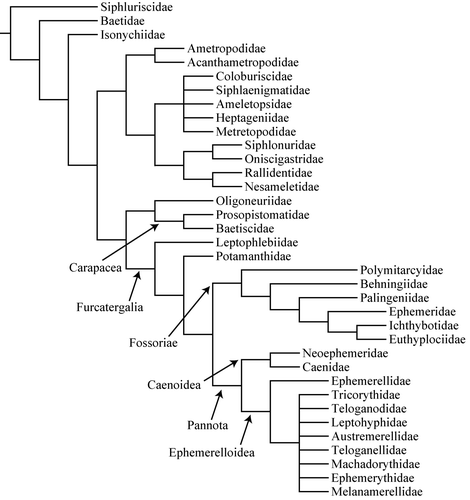
The higher classification of mayflies based on morphology and anatomy was presented independently by McCafferty (Citation1991) with subsequent revisions (e.g. McCafferty and Wang Citation2000; Wang and McCafferty 2004; Jacobus and McCafferty Citation2006), and by Kluge (Citation2004). Both reconstructions are congruent at a number of deep nodes, including the basal position of the suborder Carapacea (or Posteritorna) and monophyly of Furcatergalia, Setisura and Siphlonuroidea. The first molecular phylogeny of the mayflies, using 31 of 37 families and nearly one-quarter of all genera (Ogden and Whiting Citation2005) recovered the Furcatergalia and Carapacea as monophyletic, but not Setisura or Pisciforma. Baetidae was the sister to all other mayfly clades, and a ‘fishlike’ body form was plesiomorphic and subject to several secondary losses.
More recently, a combined analysis (; Ogden et al. 2009) of more than 100 morphological characters and 5800 bp of mitochondrial (12S, 16S) and nuclear (18S, 28S, H3) genes has generated several interesting new insights. First, Carapacea is never recovered as the basal group of mayflies but is nested together with the family Oligoneuriidae as the sister group of Furcatergalia. This suggests that the posterior condition of the anal field in the forewing is not plesiomorphic. The most basal family is Siphluriscidae, which confirms its ancient origin as has been suggested (Zhou and Peters Citation2003), followed by the Baetidae and Isonychiidae. Neither Setisura nor Siphlonuroidea were recovered as monophyletic and relationships among them are still unclear. Among Furcatergalia which is recovered as monophyletic, the Fossoriae sensu Kluge (Citation2004) forms a monophyletic group with Pannota as sister group. One of the key contributions of this work was to demonstrate the high degree of homoplasy in the morphological characters that were used. A molecular phylogenetic study of several genera of Baetidae also found repeated switches in mouthpart morphology within single lineages (Monaghan et al. Citation2005). These results strongly suggest that morphological characters will provide a misleading picture of mayfly phylogeny.
Biogeography and family-level phylogenetics
Within families, two molecular phylogenies are available that provide important information on mayfly biogeography. One is a global scale phylogeny for the Leptophlebiidae and one an Afrotropical phylogeny of the Baetidae. We note that these are the most species- and genus- rich families of mayflies (Barber-James et al. Citation2008) but that virtually nothing is known of the remaining 40 families. In a study of 30 genera (plus 11 taxa of unknown rank) of Leptophlebiidae, the recognised subfamilies Leptophlebiinae and Habrophlebiinae were recovered as monophyletic groups (O'Donnell and Jockusch Citation2008). The third and largest subfamily, Atalophlebiinae, was monophyletic only under certain tree-search parameters. In most reconstructions it was paraphyletic with respect to either Leptophlebiinae or with the Ephemerellidae. Four well-supported clades were consistently recovered in the Atalophlebiinae, one in Madagascar; one in Madagascar and New Guinea; one in Madagascar, southern South America, southern Africa, and Australia-New Zealand; and one widespread lineage. The tree of O'Donnell and Jockusch (Citation2008) now provides the framework for more detailed study of character evolution within this species-rich group.
The phylogenetic study of 26 genera of Afrotropical Baetidae was primarily aimed at establishing the origin and evolutionary depth of Malagasy lineages (Monaghan et al. Citation2005). Monophyly of the subfamilies Baetinae and Cloeoninae was moderately supported, and the analysis recovered seven well-supported lineages with members in Madagascar. Only one was entirely endemic, while four were Afrotropical, and two included Asian and European taxa. Thus the Malagasy taxa appear to be part of a much larger spatial context, having undergone repeated exchange with other landmasses. This finding is similar to that of Leptophlebiidae, with three distinct lineages within the Atalophlebiinae (above). Ancestral state likelihood analysis suggested the genus Cloeodes had its origin in Madagascar, and that a scenario of unidirectional dispersal from a continental source is too simplistic (Monaghan et al. Citation2005). This was somewhat surprising because nominal Cloeodes has a Gondwanian distribution; however, preliminary genetic analysis indicates South American and Afrotropical Cloeodes are unrelated (C. Nieto and M.T. Monaghan, unpublished).
Basal phylogeny of the winged insects (Pterygota)
Until the recent progress summarised above, the phylogenetic study of mayflies has been focused on determining their position within the insects as a whole. In most studies, one or a few species of mayflies are chosen to represent the Order. In particular, the phylogenetic position of the mayflies within the Pterygota has been a subject of interest, with three competing hypotheses. The Paleoptera hypothesis states that Ephemeroptera + Odonata are sister to Neoptera, with alternatives being a basal Ephemeroptera hypothesis (Ephemeroptera + (Odonata + Neoptera)) and a basal Odonata hypothesis (Odonata + (Ephemeroptera + Neoptera)). Each hypothesis has support from morphological characters (see Ogden and Whiting Citation2003; Zhang et al. Citation2008), and molecular phylogenetics has been employed with the hope of providing independent support for one of these.
In some of the more recent attempts to address the relationships among basal pterygotes, Hovmöller et al. (Citation2002) analysed two nuclear genes, 18S and 28S rRNA, from the ribosomal subunit, and found evidence for the Paleoptera hypothesis. Ogden and Whiting (Citation2003) found that the results of basal pterygote reconstructions were highly sensitive to alignments and to the method of tree reconstruction. Even with the addition of a third gene (protein-coding histone 3), they found evidence for both the Paleoptera and Ephemeroptera hypotheses depending on alternate alignment methods. Using a broader taxon sample and only the ribosomal subunit data (18S, 28S), Mallatt and Giribet (Citation2006) found evidence for the Odonata hypothesis. Finally, a supermatrix approach of all available data, combining four nuclear and five mitochondrial genes with 170 morphological characters supported the Paleoptera hypothesis (Kjer et al. Citation2006a). Thus it could be argued that the debate has come full circle.
An exciting development in the study of basal Pterygota was provided by the first complete mitochondrial genome of a mayfly, that of the heptageniid Parafronurus youi (Zhang et al. Citation2008). Among several notable findings from their study was that Ephemeroptera was a well supported sister taxon to Odonata + Neoptera in a mitochondrial phylogeny of nine insect orders. Several of the findings of Zhang et al. (Citation2008) were in agreement with what is the largest mitochondrial phylogeny of arthropods that is presently available (Carapelli et al. Citation2007), however, the mayfly data were not yet published and thus Carapelli et al. (Citation2007) could not yet test any of the Ephemeroptera-Odonata-Neoptera hypotheses. There will certainly be more to come in the area of mitochondrial genomics.
The work to date has provided some important advances and has identified important areas for future work. While molecular studies have yet to reach a consensus, the topic has generated a healthy debate regarding the gene regions and the phylogenetic methods employed (e.g. Ogden and Whiting Citation2003; Kjer et al. Citation2006b). That the analyses and genetic markers used to date fail to strongly support any hypothesis to the exclusion of the other stems, in part, from the limitations of the data themselves. Most studies have considered only one or two gene regions, namely 18S and 28S, both of which are part of the ribosomal subunit. This is largely because of the practical difficulty of collecting sequence data across multiple markers from a broad range of taxa. Although rRNA markers are highly variable and comparatively easy to amplify across taxa, their length variation (i.e. presence of insertion/deletion events) can be problematic. As discussed above, small changes to alignment parameters and tree reconstruction methods can yield conflicting results. Protein-coding genes minimise the difficulties of alignment, and homoplasy in nucleotide or amino acid evolution can be effectively modelled using likelihood and Bayesian approaches. One limitation of protein-coding regions can be that constraints on gene function lead to their being highly conserved. This can mean they contain limited phylogenetic signal. This was argued to be the case for h3 by Kjer et al. (Citation2006a) and appears to be supported by the results of Ogden and Whiting (Citation2003). The need for more nuclear markers in mayfly research is critical, and is discussed further in the Future perspectives section.
Evolutionary implications of the position of Ephemeroptera
Resolving the basal relationships of the Pterygota (above) represents a critical step toward understanding the early diversification of the insects that now form the large diversity of eukaryotic life on Earth. Certainly the early changes in reproduction, life history, and flight have played an important role in generating this biodiversity. Here we highlight several findings from other avenues of genetics research that underline the importance of clarifying the position of mayflies. Mayflies have distinct characteristics at the whole-genome scale that, because of their basal position, could shed additional light on insect evolution.
The ends of chromosomes (telomeres) in most eukaryotes are characterised by long stretches of short tandem repeats in the DNA sequence. These play an important role in telomere elongation during chromosome replication. TTAGG telomeric repeats are ancestral in the arthropods but have been lost in the Paleoptera (Frydrychova et al. Citation2004). The fact that the repeats have also been lost several times in Diptera and Coleoptera suggests the TTAGG telomeric repeats could have important implications for the evolution and diversification of some of the most species-rich groups of insects (Frydrychova and Marec Citation2002).
Katayama (Citation1939) established that males are the heterogametic sex (XY) in Ameletus costalis and this was supported by Wolf (Citation1960) and Kiauta and Mol (Citation1977) in Cloeon. Soldán and Putz (Citation2000) found two types of heterogametic males, XY in Baetidae, Heptageniidae, Ephemerellidae, Oligoneuriidae, and Siphlonuridae; and X0 in Ephemeridae, Potamanthidae, and Caenidae (i.e. a second sex chromosome was absent in males). Because Odonata have X0 determination, it would appear from a cursory placement of characters on the phylogenetic tree of Ogden and Whiting (Citation2005) that XY determination arose early in the mayflies and reverted to X0 at least once within the suborder Furcatergalia. It would be very interesting to examine sex determination in the recently discovered Siphluriscidae (Zhou and Peters Citation2003) which appears to be the evolutionary link between Odonata and Ephemeroptera ().
More recently, a notable finding from the mitochondrial genome sequence discussed earlier was that the origin of replication (“AT-rich region”) in mayflies has the lowest known A + T content of any extant hexapod (Zhang et al. Citation2008). There is also evidence that mayflies have one of the smallest genomes of the hemimetabolous insects (Gregory Citation2005). The small size is similar to that of many holometabolous insects. Among several competing hypotheses of insect genome size is that smaller genomes more readily allow for complete metamorphosis. Certainly an intriguing possibility is that mayflies, which alone retain the subimago stage, provide a critical link in the evolution of genome size and the transition from incomplete to complete metamorphosis.
Population genetics
Population genetics can be thought of as the study of genetic variation within species, although journals and articles that cover population genetics are increasingly interested in genetic variation at the population-species boundary. Here we consider the works published on mayflies that reported genetic variation within species. In most cases, authors were primarily interested in heterozygosity, or genetic diversity per se, and in using genetics to estimate levels of dispersal among populations in geographical space.
Heterozygosity
Under conditions of equilibrium (i.e. with random mating and no selection, genetic drift, or mutation), the number of individuals with two different copies (alleles) of a given gene, or heterozygotes, can be predicted from the frequency of alleles in the population. This calculation uses the Hardy–Weinberg equation. When the measure of heterozygous individuals is different than the expected number from the allele frequency data, one of the conditions is violated. An early study of genetic variation in populations of Baetis harrisoni found alleles of the allozyme locus Pgm to be in Hardy–Weinberg equilibrium (Agnew Citation1978). The majority of studies since then have used multiple loci and the large majority have found lower than expected heterozygosities in mayfly populations (). Low heterozygosity, also referred to as inbreeding, has been observed in mayflies from Australia, Europe, New Zealand and North America, and a number of hypotheses have been put forth in explanation.
Table 1. Population genetics studies of mayflies (Ephemeroptera), here defined as publications that reported F ST based on at least two populations of a single (nominal) species.
The ‘patchy recruitment hypothesis’ (Schmidt et al. Citation1995; Bunn and Hughes Citation1997) states that populations are founded by small number of females in each generation. This constitutes a bottleneck and leads to low heterozygosities. The hypothesis has received the most attention and subsequent testing, with mixed results. Studies that combine mtDNA and allozymes (see below) report a small number of mtDNA haplotypes. Because mtDNA is transmitted maternally, this can be interpreted as evidence for populations being founded by a small number of females. Other explanations for low heterozygosity include natural selection and automictic parthenogenesis. There is strong evidence in the literature for natural selection favouring alleles at some loci in some cases, although this has not been explicitly tested for mayflies. But most of the reduction of heterozygosity in mayflies appears random, rather than at the same loci as might be expected if loci were under selection (Bunn and Hughes Citation1997). Parthenogenesis has been documented in a number of mayfly species (e.g. Needham Citation1924; Degrange Citation1954; Gibbs Citation1977; Sweeney and Vannote Citation1987). Apomictic parthenogenesis is expected to increase heterozygosity in populations because new mutations generally occur as heterozygotes and loci are expected to remain heterozygous in the absence of recombination. Interestingly, Centroptilum triangulifer undergoes automictic parthenogenesis which can reduce heterozygosity, although in this case there was also an excess of heterozygotes (Funk et al. Citation2006). We would benefit from a greater understanding of whether the populations and species examined at the population level to date () are parthenogenetic, but there is little evidence to date to suggest that automictic parthenogenesis contributes to low heterozyosities in natural populations.
Wilcock et al. (Citation2005) explicitly tested the patchy recruitment hypothesis in caddisflies by measuring oviposition and genetic relatedness of populations of Plectrocnemia conspersa. They were able to identify groups of siblings within egg masses, but found that larvae dispersed rapidly and sib-group structure was not retained. Our understanding of low heterozygosity in mayflies would benefit greatly from similar experiments that combine genetic tools and field experiments for a more mechanistic understanding.
Ecology and evolution
A number of ecological and evolutionary hypotheses have been explicitly examined using genetic methods. Peckarsky et al. (Citation2005) tested whether nocturnal behaviour and accelerated development of Baetis larvae in fish streams was an example of phenotypic plasticity or whether populations were genetically different from nearby fishless streams. They found no genetic differences associated with fish occurrence, a finding supported by experimental evidence that fish chemical cues can initiate such behaviour in Baetis originating from fishless streams (e.g. Peckarsky et al. Citation2002). Robinson et al. (Citation1992) tested whether mayfly populations living in more variable habitats had greater genetic variation than those in more stable habitats. Comparing a river with discharge dominated by groundwater with a river dominated by snowmelt, they found increased heterozygosity in the more variable, snowmelt-fed river (Robinson et al. Citation1992). Their findings fit with patterns observed in many other taxa, that the local environmental stability can have important effects on genetic variation (e.g. Hedrick Citation1986). This was the case even in the presence of what appeared to be regular gene flow between the two populations.
Dispersal
The study of dispersal has been a primary focus of population genetics research on mayflies. With spatially explicit allele frequency data, population genetic models can be used to infer population connectedness i.e. the magnitude of gene flow within a species. Seventeen species have been examined to date, using allozymes, microsatellites or mtDNA. Nearly all studies report significant departure from panmixia at scales of 1–1500 km, and frequently at scales <100 km (). Perhaps more significant is that the extent of differentiation varies more than 30-fold, from 0.007 < F st < 0.210. This kind of variation makes it difficult to generalise, and indeed the sizeable variation among species is one of the major conclusions of population genetics research. Sweeney et al. (Citation1986) studied two ephemerellids that co-occur in the same drainage system. Ephemerella subvaria exhibited significant differentiation among sites within and among tributaries of the same river basin (ca. 10–100 km), whereas Eurylophella verisimilis exhibited near panmixia at the same sites. Monaghan et al. (Citation2002) examined two mayfly species with very similar distributions in high Alpine streams, and yet found a threefold difference in gene flow of Rhithrogena loyolaea compared to Baetis alpinus, the former seemingly differentiated only across major drainage barriers. Similarly, genetic differentiation within populations of two co-occurring species of Baetis in the North American Rockies differed by nearly an order of magnitude over scales of 1–20 km (Peckarsky et al. Citation2005).
Three mayfly species have been examined using both allozymes and mtDNA. These are Bungona narilla in Australia, Baetis bicaudatus in North America and Acanthophlebia cruentata in New Zealand (). A very consistent pattern is that genetic differentiation among populations is higher with mtDNA than allozymes. Two hypotheses have been proposed in explanation. First, the mitochondrial genome is haploid and maternally inherited, meaning its effective population size is one-quarter that of nuclear genes. This makes it more sensitive to bottlenecks, colonisation events, and stochastic genetic drift (e.g. Hughes et al. Citation2003b; Smith et al. Citation2006). Alternatively, greater population distinctiveness in females could be evidence for male-biased dispersal (e.g. Hughes et al. Citation2003b). In support of the latter, Petersen et al. (Citation2004) found that male mayflies (Baetis, Ephemerella, Heptagenia) were more abundant in riparian areas and travelled farther from the stream. In contrast, Caudill (Citation2003) found evidence for female-biased dispersal among ponds in Callibaetis using stable isotopes.
Hughes (Citation2007) pointed out that genetic studies have contributed to a view of dispersal and recolonisation that includes broader spatial scales. It is quite clear that the spatial extent at which stream ecologists study recolonisation has increased substantially over the past decades; compare the reviews by Mackay (1995) and Covich (Citation2005). Recent studies of dispersal and colonisation among streams have interpreted their results in part based on gene flow studies. Interestingly, both concluded that dispersal among streams was not a limiting factor, based on finding adult mayflies near polluted streams with no larval populations (Masters et al. Citation2007) or inferred from patterns of distribution and nestedness (Monaghan et al. Citation2005).
Two studies examined how reservoirs might disrupt gene flow between populations living only in free-flowing reaches of rivers and streams. Sweeney et al. (Citation1986) first examined the effect of reservoirs on mayfly dispersal and found no evidence for disruption of gene flow. Monaghan et al. (Citation2001, Citation2002) observed similar patterns in Alpine streams and concluded that any genetic effects of reservoirs are probably too recent to identify with allozyme markers. Differentiation across reservoirs has been observed more recently in a caddisfly using RAPD methods to analyse 52 loci (Watanabe and Omura Citation2007), suggesting that studies targeting more variable parts of the genome, or assaying a larger number of polymorphic loci might identify more recent genetic divergence.
A first conclusion that we can draw from studies to date is that results vary widely among species, even closely related congeners, and that it can be difficult and misleading to make general statements about mayfly dispersal. This can be seen clearly in our summary of differentiation values (). Once this variation is taken into account, a second conclusion is that mayflies probably disperse more widely than expected based on field studies. For example, Petersen et al. (Citation2004) found that 50% of the adult mayflies captured travelled less than 17 m and 90% travelled less than 59 m. From these data we might conclude that dispersal among streams is limited and that the genetics data contradict such findings. But it is important to remember that a strength of genetic methods is their capacity to capture the rare but potentially important events. Many studies from the large literature on dispersal find at least one individual in the most distant trap, and these may be the critical events. According to population genetics theory, only low levels of migration are necessary to avoid fixation of alternate alleles, thus these rare long-distance events can make all the difference to population genetic signatures.
A third conclusion is that the temporal scales of resolution for population genetic data have yet to be fully established. It is difficult to determine whether an observed level of differentiation between populations results from an equilibrium between genetic drift and gene flow, or if it is the result of population history, having failed to reach an equilibrium among the newly subdivided populations (Broughton and Harrison Citation2003). Several studies have concluded that genetic patterns result from a combination of contemporary and historical gene flow (e.g. Smith and Collier Citation2001; Monaghan et al. Citation2002). As mentioned earlier, what may appear to be sustained gene flow across reservoirs may in reality be older genetic signals from markers (Monaghan et al. Citation2001). Population genetics infers gene flow from random changes in allele frequency and the eventual fixation of alternate alleles by genetic drift. This is a function of time and population size, and over short timescales and for large populations these differences may not be detectable. In the final section of this paper we discuss some promising advances in this field, such as the use of multiple markers and the testing of explicit hypotheses about separation times, but these techniques have yet to be applied to mayflies.
Future perspectives
Taxonomy
Species delineation continues to be one of the primary applications of genetic techniques, some 24 years after the original attempts. As we argued above, technology alone (i.e. DNA sequencing) has done little to circumvent the same problems faced by those original researchers. An area of research that does look very promising is the application of the generalised mixed Yule-coalescent (GMYC) model to species circumscription using single-locus DNA (Pons et al. Citation2006; Fontaneto et al. Citation2007). The GMYC attempts to identify the species-population boundary as a shift in branching rates on a tree that contains DNA sequences from multiple species and populations. Branching patterns reflect either neutral coalescent processes occurring within species or speciation events (Pons et al. Citation2006). The GMYC assesses the point of highest likelihood of the transition, and independently evolving lineages are recovered as putative species, more distinct than predicted if the entire sample was derived from a single species without genetic isolation. The approach has been applied successfully to 64 species of mayfly in Madagascar (Monaghan et al. 2009) with an important consequence being the ability to establish the necessary sample sizes of between 7 and 10 individuals per putative species (see also Morando et al. Citation2003). Most studies using DNA sequence variation in taxonomy provide little justification of sample sizes employed (Ball et al. Citation2005; Savolainen et al. Citation2007; Webb et al. Citation2007). While samples of some groups may be limited by their rarity in the field, it is important to consider how small sample sizes may affect any conclusions of group membership in the absence of additional data.
Population genetics
The mayflies currently comprise more than 3000 recognised species within 42 families (Barber-James et al. Citation2008), but there are only population-level studies for 17 species, 12 of which were either Baetidae or Ephemerellidae (). Thus, the great diversity of mayfly lineages is poorly represented or completely lacking. It is also important to consider that much of the existing work has been based on samples from larval populations. This is partly because the long residence time in the water as immatures makes them important ecologically, makes them suitable for biomonitoring, and makes large numbers easier to sample. It may also result from the relatively small amount of tissue available from adult specimens, although a protocol for obtaining high molecular weight DNA from adult males has been developed (Takemon et al. Citation2006) that may prove very useful for the genetic studies of adult life stages.
Resolving the temporal scales at which genetic markers reveal patterns remains one of the critical tasks for population geneticists. Studies of gene flow often begin with the intention of investigating contemporary patterns of gene flow but find evidence that what the data reveal are longer-term patterns. Hierarchical sampling is one means of addressing the problem, whereby larger spatial scales should reflect older patterns, and smaller scales the more recent patterns (e.g. Hellberg Citation1994). Nested clade analysis (NCA) provides, in principle, a means to infer how contemporary and historical gene flow interact to generate patterns and has been employed in mayfly research (e.g. Smith et al. Citation2006; McLean et al. Citation2008). Unfortunately, NCA is increasingly criticised, primarily because the conditions under which inferences are accurate have yet to be identified (Knowles Citation2008).
More recent methods of genetic analysis that employ coalescence potentially provide a better temporal perspective on genetic processes. Investigators of the demographic history of closely related populations or species can use several nuclear DNA sequences to test specific hypotheses of how past geological events influence observed patterns (e.g. Knowles et al. Citation2007). These techniques allow one to test a priori hypotheses rather than draw post hoc conclusions from patterns. A good understanding of the rate of sequence evolution is critical to interpreting some of these analyses; however, rates are poorly known. Most studies that employ an explicit rate of sequence evolution that continue to use that of Brower's (Citation1994) study of Heliconius butterflies. Finally, the low A + T value of the AT-rich region in mayfly mtDNA (Zhang et al. Citation2008) suggests the region could be more easily amplified and sequenced than in other insects. This could provide a highly variable molecular marker for population-genetic (e.g. Schultheis et al. Citation2002) and species-level studies using coalescent approaches.
New technologies
As of early October 2008 there were 1455 Ephemeroptera sequences on the nucleotide database of GenBank. For comparison, there were 1088 Plecoptera, 2104 Trichoptera, 3602 Odonata, 1695 Chironomidae, and 1,167,737 Drosophila. These values were all recovered using the above search term and filtering with the tree of ‘Top Organisms’ using the taxonomy database. While the differences seem daunting, next-generation sequencing technologies (e.g. ‘pyrosequencing’) provide the means to equate the number of mayfly sequences with that of Drosophila in only a few days. By the time this article is in print, it may be possible within one day. Research projects are already limited by the rate of statistical analysis rather than data generation. As we argued with species circumscription, data alone are of limited value without clear concepts and testable hypotheses. An important consequence is that the limiting factor in many phylogenetics studies will be sample availability from under-represented areas of the globe, which should be brought to the attention of funding agencies and permit-issuing authorities.
Increased sequencing technology could soon make museum collections more widely accessible to sequencing reactions. Museum collections provide a wealth of genetic material. This is often of historical importance, such as type specimens or extinct species or populations. Most genetic information in museums remains unavailable for study because current sequencing techniques rely on high-quality DNA. Museum preservation techniques (e.g. 70% ethanol) are designed for morphological analysis but this is destructive to DNA, breaking it into small pieces that are difficult or impossible to amplify using standard PCR. Pyrosequencing of these short DNA fragments using ‘sequencing-by-synthesis’ provides an exciting tool to examine degraded DNA (e.g. Poinar et al. Citation2007). This will open new research avenues for collection-based genetic research, and increase the scientific and cultural value of museum collections. The integration of collections and pyrosequencing would provide a taxonomic scaffold of type specimens for the use of DNA in species description and identification.
Twice in our review we have identified significant advances that would come about with the availability of sequences of more nuclear markers. Not only would these help phylogenetics (currently limited by rRNA and h3 data), but multiple nuclear markers are likely to soon enhance the power of single-locus mtDNA as a means of species circumscription and more detailed studies of speciation. Based on their phylogenetic position, it is somewhat surprising that a complete mayfly genome is unavailable, but as we have seen it is comparatively recently that the first mtDNA genome for a mayfly was published.
Conclusions
Species delineation, population genetics, and phylogenetics have been treated separately in our review, but the relative ease of collecting genetic data from a broad range of species using conserved primers means that the areas of study increasingly overlap. Most of the studies examining genetic variation within species have focused on one or two species, although Zurwerra et al. (Citation1987) examined 55 taxa from as many as eight populations, and Sweeney et al. (Citation1987) examined 10 species from as many as 16 populations. More recent studies typically examine genetic variation within one or more lineages rather than a priori species (e.g. Williams et al. Citation2006; Stahls and Savolainen Citation2008; Pereira da Conceicoa Citation2008; Monaghan et al. 2009) and this certainly is an evolutionarily realistic approach to understanding biodiversity. More importantly, increasing use of coalescence and phylogenetics analyses is linking population-genetic and phylogenetic disciplines in exciting ways. But while taxonomy, biogeography, speciation, and evolution will be increasingly understood using these approaches, it is important to remember that some of the most informative studies we review here have used genetic tools as one of many components for testing specific hypotheses of mayfly biology. Examples include measuring genetic variation in differentially disturbed habitats (Robinson et al. Citation1992), phenotypic plasticity in response to the presence of fish predators (Peckarsky et al. Citation2005), and parthenogenetic reproduction (Funk et al. Citation2006). Wilcock et al. (Citation2005) demonstrated very well how the combination of ecology and genetics research, applied to several parts of the life cycle, can greatly advance our understanding of how populations function in nature. It is our hope that the above review has highlighted some of these advances and will encourage other workers to pursue these and other topics of research in mayflies.
Acknowledgements
We extend our gratitude to Arnold Staniczek and the staff at the Staatliches Museum für Naturkunde in Stuttgart for hosting the International Joint Meeting on Ephemeroptera and Plecoptera 2008. Michael Hubbard maintains an outstanding bibliography of mayfly research at Ephemeroptera Galactica ( http://www.famu.org/mayfly/mfbib.php) and Helen Barber-James provided several references. We appreciate the fruitful conversations with many conference attendees, as well as comments on the manuscript by Bern Sweeney and one anonymous referee. Our research on mayfly genetics has been funded in part by the UK BBSRC (BBS/B/04358) and the Swiss National Fund (Stipendium Nr. 68592, Research Grant Nr. 3100A0-116049).
References
- Agnew , J. D. ‘The Pgm locus in Baetis harrisoni’ . Proceedings of the 6th Congress of the South African Genetics Society . pp. 180 – 183 . Pretoria : South African Genetics Society .
- Ball , S. L. , Hebert , P. D.N. , Burian , S. K. and Webb , J. M. 2005 . ‘Biological identifications of mayflies (Ephemeroptera) using DNA barcodes’ . Journal of the North American Benthological Society , 24 : 508 – 524 .
- Barber-James , H. M. , Gattolliat , J.-L. , Sartori , M. and Hubbard , M. D. 2008 . ‘Global diversity of mayflies (Ephemeroptera, Insecta) in freshwater’ . Hydrobiologia , 595 : 339 – 350 .
- Bazin , E. , Glemin , S. and Galtier , N. 2006 . ‘Population size does not influence mitochondrial genetic diversity in animals’ . Science , 312 : 570 – 572 .
- Belfiore , C. , Picariello , O. , Scillitani , G. and Cretella , M. 1992 . ‘Genetic divergence between two insular species of the genus Rhithrogena (Ephemeroptera, Heptageniidae)’ . Fragmenta Entomologica , 23 : 235 – 242 .
- Belfiore , C. , Scillitani , G. , Picariello , O. and Cataudo , A. 1997 . ‘Morphological and electrophoretic evidence for a new species of Electrogena from Central Italy: description of E. lunaris sp. n.’ . Aquatic Insects , 19 : 129 – 140 .
- Broughton , R. E. and Harrison , R. G. 2003 . ‘Nuclear gene genealogies reveal historical, demographic and selective factors associated with speciation in field crickets’ . Genetics , 163 : 1389 – 1401 .
- Brower , A. V.Z. 1994 . ‘Rapid morphological radiation and convergence among races of the butterfly Helioconius erato inferred from patterns of mitochondrial DNA evolution’ . Proceedings of the National Academy of Sciences , 91 : 6491 – 6495 .
- Bunn , S. E. and Hughes , J. M. 1997 . ‘Dispersal and recruitment in streams: evidence from genetic studies’ . Journal of the North American Benthological Society , 16 : 338 – 346 .
- Carapelli , A. , Liò , P. , Nardi , F. , van der Wath , E. and Frati , F. 2007 . ‘Phylogenetic analysis of mitochondrial protein coding genes confirms the reciprocal paraphyly of Hexapoda and Crustacea’ . BMC Evolutionary Biology , 7, Supplement 2 : S8
- Caudill , C. C. 2003 . ‘Measuring dispersal in a metapopulation using stable isotope enrichment: high rates of sex-biased dispersal between patches in a mayfly metapopulation’ . Oikos , 101 : 624 – 630 .
- Covich , A. P. 2005 . ‘Dispersal-limited biodiversity of tropical insular streams’ . Polish Journal of Ecology , 54 : 523 – 547 .
- Davis , J. I. and Nixon , K. C. 1992 . ‘Populations, genetic variation, and the delimitation of phylogenetic species’ . Systematic Biology , 41 : 421 – 435 .
- Degrange , M. C. 1954 . ‘Deux cas de parthénogenèse chez les Ephéméroptères: Siphlonurus aestivalis Eat et Centroptilum luteolum Müll’ . Comptes Rendus des Séances de l'Académie des Sciences , 239 : 1082 – 1083 .
- Felsenstein , J. 2004 . Inferring Phylogenies , Sunderland , MA : Sinauer .
- Fontaneto , D. , Herniou , E. A. , Boschetti , C. , Caprioli , M. , Melone , G. , Ricci , C. and Barraclough , T. G. 2007 . ‘Independently evolving species in asexual bdelloid rotifers’ . PLoS Biology , 5 : e87
- Forney , L. J. , Zhou , X. and Brown , C. J. 2004 . ‘Molecular microbial ecology: land of the one-eyed king’ . Current Opinion in Microbiology , 7 : 210 – 220 .
- Frydrychova , R. , Grossmann , P. , Trubac , P. , Vitkova , M. and Marec , F. E. 2004 . ‘Phylogenetic distribution of TTAGG telomeric repeats in insects’ . Genome , 47 : 163 – 178 .
- Frydrychova , R. and Marec , F. E. 2002 . ‘Repeated losses of TTAGG telomere repeats in evolution of beetles (Coleoptera)’ . Genetica , 115 : 179 – 187 .
- Funk , D. H. , Jackson , J. K. and Sweeney , B. W. 2006 . ‘Taxonomy and genetics of the parthenogenetic mayfly Centroptilum triangulifer and its sexual sister Centroptilum alamance (Ephemeroptera:Baetidae)’ . Journal of the North American Benthological Society , 25 : 417 – 429 .
- Funk , D. H. , Sweeney , B. W. and Jackson , J. K. 2008 . ‘A taxonomic reassessment of the Drunella lata (Morgan) species complex (Ephemeroptera : Ephemerellidae) in northeastern North America’ . Journal of the North American Benthological Society , 27 : 647 – 663 .
- Gibbs , H. L. , Gibbs , K. E. , Siebenmann , M. and Collins , L. 1998 . ‘Genetic differentiation among populations of the rare mayfly Siphlonisca aerodromia Needham’ . Journal of the North American Benthological Society , 17 : 464 – 474 .
- Gibbs , K. E. 1977 . ‘Evidence for obligatory parthenogenesis and its possible effect on the emergence period of Cloeon triangulifer (Ephemeroptera: Baetidae)’ . Canadian Entomologist , 109 : 337 – 340 .
- Godfray , H. C.J. 2002 . ‘Challenges for taxonomy’ . Nature , 417 : 17 – 19 .
- Gregory , T. R. 2005 . “ ‘Genome size evolution in animals’ ” . In The Evolution of the Genome , Edited by: Gregory , T. R. 3 – 87 . San Diego : Elsevier .
- Hartl , D. L. and Clark , A. G. 1997 . Principles of Population Genetics, 3rd ed , Sunderland , MA : Sinauer .
- Hedrick , P. W. 1986 . ‘Genetic polymorphism in heterogeneous environments: a decade later’ . Annual Review of Ecology and Systematics , 17 : 535 – 566 .
- Hellberg , M. E. 1994 . ‘Relationships between inferred levels of gene flow and geographic distance in a philopatric coral, Balanophyllia elegans’ . Evolution , 48 : 1829 – 1854 .
- Hogg , I. D. , Willmann-Huerner , P. and Stevens , M. I. 2002 . ‘Population genetic structures of two New Zealand stream insects: Archichauliodes diversus (Megaloptera) and Coloburiscus humeralis (Ephemeroptera)’ . New Zealand Journal of Marine and Freshwater Research , 36 : 491 – 501 .
- Hovmöller , R. , Pape , T. and Källersjö , M. 2002 . ‘The Palaeoptera problem: basal pterygote phylogeny inferred from 18S and 28S rDNA sequences . Cladistics , 18 : 313 – 323 .
- Hughes , J. M. 2007 . ‘Constraints on recovery: using molecular methods to study connectivity of aquatic biota in rivers and streams’ . Freshwater Biology , 52 : 616 – 631 .
- Hughes , J. M. , Hillyer , M. and Bunn , S. E. 2003a . ‘Small-scale patterns of genetic variation in the mayfly Bungona narilla (Ephemeroptera: Baetidae) in rainforest streams, south-east Queensland’ . Freshwater Biology , 48 : 709 – 717 .
- Hughes , J. M. , Mather , P. B. , Hillyer , M. J. , Cleary , C. and Peckarsky , B. 2003b . ‘Genetic structure in a montane mayfly Baetis bicaudatus (Ephemeroptera: Baetidae), from the Rocky Mountains, Colorado . Freshwater Biology , 48 : 2149 – 2162 .
- Jacobus , L. M. and McCafferty , W. P. 2006 . ‘Reevaluation of the phylogeny of the Ephemeroptera Infraorder Pannota (Furcatergalia), with adjustments to higher classification’ . Transactions of the American Entomological Society , 132 : 81 – 90 . 429–430
- Katayama , H. 1939 . ‘The sex chromosomes of a May-fly, Ameletus costalis Mats. (Ephemerida)’ . Japanese Journal of Genetics , 15 : 139 – 144 .
- Kiauta , B. and Mol , A. W.M. 1977 . ‘Behaviour of the spermatocyte chromosomes of the mayfly, Cloeon dipterum (Linnaeus, 1761) s.l. (Ephemeroptera: Baetidae), with a note on the cytology of the order’ . Genen en Phaenen , 19 : 31 – 39 .
- Kjer , K. M. , Carle , F. L. , Litman , J. and Ware , J. 2006a . ‘A molecular phylogeny of Hexapoda’ . Arthropod Systematics and Phylogeny , 64 : 3 – 44 .
- Kjer , K. , Gillespie , J. J. and Ober , K. A. 2006b . ‘Structural homology in ribosomal RNA, and a deliberation on POY’ . Arthropod Systematics and Phylogeny , 64 : 159 – 164 .
- Kluge , N. 2004 . The Phylogenetic System of Ephemeroptera , Dordrecht : Kluwer .
- Knowles , L. L. 2008 . ‘Why does a method that fails continue to be used?’ . Evolution , 62 : 2713 – 2717 .
- Knowles , L. L. , Carstens , B. C. and Keat , M. L. 2007 . ‘Coupling genetic and ecological-niche models to examine how past population distributions contribute to divergence’ . Current Biology , 17 : 940 – 946 .
- Kownacki , A. and Starmach , J. ‘Electrophoretic investigations on taxonomy of some species of Ephemeroptera from the Dunajec basin (Poland)’ . Proceedings of the Fourth International Conference on Ephemeroptera . Edited by: Landa , V. , Soldán , T. and Tonner , M. pp. 219 – 224 . České Budějovice : Institute of Entomology, Czechoslovak Academy of Sciences .
- Landolt , P. 1991 . “ ‘An approach to the application of molecular biological methods to solve taxonomical and phylogenetic problems of the Ephemeroptera’ ” . In Overview and strategies of Ephemeroptera and Plecoptera , Edited by: Alba-Tercedor , J. and Sánchez-Ortega , A. 3 – 14 . Gainesville , FL : Sandhill Crane Press .
- Mackay , R. J. 1992 . ‘Colonization by lotic macroinvertebrates: a review of processes and patterns’ . Canadian Journal of Fisheries and Aquatic Sciences , 49 : 617 – 628 .
- Mallatt , J. and Giribet , G. 2006 . ‘Further use of nearly complete 28S and 18S rRNA genes to classify Ecdysozoa: 37 more arthropods and a kinorhynch’ . Molecular Phylogenetics and Evolution , 40 : 772 – 794 .
- Masters , Z. , Petersen , I. , Hildrew , A. G. and Ormerod , S. J. 2007 . ‘Insect dispersal does not limit the biological recovery of streams from acidification’ . Aquatic Conservation: Marine and Freshwater Ecosystems , 17 : 375 – 383 .
- McCafferty , W. P. 1991 . ‘Toward a phylogenetic classification of the Ephemeroptera (Insecta): A commentary on systematics’ . Annals of the Entomological Society of America , 84 : 343 – 360 .
- McCafferty , W. P. and Wang , T.-Q. 2000 . ‘Phylogenetic systematics of the major lineages of pannote mayflies (Ephemeroptera: Pannota)’ . Transactions of the American Entomological Society , 126 : 9 – 101 .
- McLean , A. J. , Schmidt , D. J. and Hughes , J. M. 2008 . ‘Do lowland habitats represent barriers to dispersal for a rainforest mayfly, Bungona narilla, in south-east Queensland?’ . Marine and Freshwater Research , 59 : 761 – 771 .
- Monaghan , M. T. , Gattolliat , J.-L. , Sartori , M. , Elouard , J-.M. , Barber-James , H. , Derleth , P. , Glaizot , O. , de Moor , F. C. and Vogler , A. P. 2005 . ‘Trans-oceanic and endemic origins of the small minnow mayflies (Ephemeroptera, Baetidae) of Madagascar’ . Proceedings of the Royal Society of London B Biological Sciences , 272 : 1829 – 1836 .
- Monaghan , M. T. , Robinson , C. T. , Spaak , P. and Ward , J. V. 2005 . ‘Macroinvertebrate species richness and nestedness in fragmented alpine streams: implications for freshwater conservation’ . Aquatic Sciences , 67 : 454 – 464 .
- Monaghan , M. T. , Spaak , P. , Robinson , C. T. and Ward , J. V. 2001 . ‘Genetic differentiation of Baetis alpinus Pictet (Ephemeroptera: Baetidae) in fragmented alpine streams’ . Heredity , 86 : 395 – 403 .
- Monaghan , M. T. , Spaak , P. , Robinson , C. T. and Ward , J. V. 2002 . ‘Population genetic structure of 3 Alpine stream insects: influences of gene flow, demographics, and habitat fragmentation’ . Journal of the North American Benthological Society , 21 : 114 – 131 .
- Monaghan , M. T. , Wild , R. , Elliot , M. , Fujisawa , T. , Balke , M. , Inward , D. J.G. , Lees , D. C. , Ranaivosolo , R. , Eggleton , P. , Barraclough , T. G. and Vogler , A. P. 2009 . ‘Accelerated species discovery on Madagascar using a coalescent-based model of species delineation’ . Systematic Biology , 58 : 298 – 311 .
- Morando , M. , Avila , L. J. and Sites , J. W. 2003 . ‘Sampling strategies for delimiting species: Genes, individuals, and populations in the Liolaemus elongatus-kriegi complex (Squamata : Liolaemidae) in Andean-Patagonian South America’ . Systematic Biology , 52 : 159 – 185 .
- Needham , J. G. 1924 . ‘The male of the parthenogenetic may-fly Ameletus ludens’ . Psyche , 31 : 308 – 310 .
- O'Donnell , B. C. and Jockusch , E. L. 2008 . ‘Phylogenetic relationships of leptophlebiid mayflies as inferred by histone H3 and 28S ribosomal DNA’ . Systematic Entomology , 33 : 651 – 667 .
- Ogden , T. H. and Whiting , M. F. 2003 . ‘The problem with “the Paleoptera problem”: sense and sensitivity’ . Cladistics , 19 : 432 – 442 .
- Ogden , T. H. and Whiting , M. F. 2005 . ‘Phylogeny of Ephemeroptera (mayflies) based on molecular evidence’ . Molecular Phylogenetics and Evolution , 37 : 625 – 643 .
- Ogden , T. H. , Gattolliat , J. L. , Sartori , M. , Staniczek , A. H. , Soldán , T. and Whiting , M. F. 2009 . ‘Toward a new paradigm in mayfly phylogeny (Ephemeroptera): combined analysis of morphological and molecular data’ . Systematic Entomology , 34 : 616 – 634 .
- Page , R. D.M. and Holmes , E. C. 1998 . Molecular evolution: a phylogenetic approach , Oxford : Blackwell Science .
- Peckarsky , B. L. , Hughes , J. M. , Mather , P. B. , Hillyer , M. and Encalada , A. C. 2005 . ‘Are populations of mayflies living in adjacent fish and fishless streams genetically differentiated?’ . Freshwater Biology , 50 : 42 – 51 .
- Peckarsky , B. L. , McIntosh , A. R. , Taylor , B. R. and Dahl , J. 2002 . ‘Predator chemicals induce changes in mayfly life history traits: a whole-stream manipulation’ . Ecology , 83 : 612 – 618 .
- Pereira da Conceicoa , L. 2008 . ‘Genetic differentiation in Baetis harrisoni (Ephemeroptera: Baetidae) in southern Africa’ unpublished honours thesis, Rhodes University, Department of Entomology and Zoology
- Petersen , I. , Masters , Z. , Hildrew , A. G. and Ormerod , S. J. 2004 . ‘Dispersal of adult aquatic insects in catchments of differing land use’ . Journal of Applied Ecology , 41 : 934 – 950 .
- Poinar , H. N. , Schwarz , C. , Qi , J. , Shapiro , B. , Macphee , R. D. , Buigues , B. , Tikhonov , A. , Huson , D. H. , Tomsho , L. P. , Auch , A. , Rampp , M. , Miller , W. and Schuster , S. C. 2007 . ‘Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA’ . Science , 311 : 392 – 394 .
- Pons , J. , Barraclough , T. G. , Gomez-Zurita , J. , Cardoso , A. , Duran , D. P. , Hazell , S. , Kamoun , S. , Sumlin , W. D. and Vogler , A. P. 2006 . ‘Sequence-based species delimitation for the DNA taxonomy of undescribed insects’ . Systematic Biology , 55 : 595 – 609 .
- Robinson , C. T. , Reed , L. M. and Minshall , G. W. 1992 . ‘Influence of flow regime on life history, production, and genetic structure of Baetis tricaudatus (Ephemeroptera) and Hesperoperla pacifica (Plecoptera) . Journal of the North American Benthological Society , 11 : 278 – 289 .
- Savolainen , E. , Drotz , M. K. , Hoffsten , P.-O. and Saura , A. 2007 . ‘The Baetis vernus group (Ephemeroptera: Baetidae) of northernmost Europe: an evidently diverse but poorly understood group of mayflies’ . Entomologica Fennica , 18 : 160 – 167 .
- Saura , A. , Lokki , J. and Savolainen , E. 1979 . ‘Ethological isolation and genetic diversity’ . Aquilo, Serie Zoologica , 20 : 13 – 16 .
- Schmidt , S. K. , Hughes , J. M. and Bunn , S. E. 1995 . ‘Gene flow among conspecific populations of Baetis sp. (Ephemeroptera): adult flight and larval drift’ . Journal of the North American Benthological Society , 14 : 147 – 157 .
- Schultheis , A. S. , Hendricks , A. C. and Weigt , L. A. 2002 . ‘Genetic evidence for ‘leaky’ cohorts in the semivoltine stonefly Peltoperla tarteri (Plecoptera: Peltoperlidae)’ . Freshwater Biology , 47 : 367 – 376 .
- Scillitani , G. , Belfiore , C. , Picariello , O. and Cataudo , A. 1996 . ‘Estimating genetic variation from larvae and adults of mayflies: an electrophoretic analysis of three species of Heptageniidae (Ephemeroptera)’ . Italian Journal of Zoology , 63 : 23 – 30 .
- Sites , J. W. and Marshall , J. C. 2004 . ‘Operational criteria for delimiting species’ . Annual Review of Ecology, Evolution, and Systematics , 35 : 199 – 227 .
- Smith , P. J. and Collier , K. J. 2001 . ‘Allozyme diversity and population genetic structure of the caddisfly Orthopsyche fimbriata and the mayfly Acanthophlebia cruentata in New Zealand streams’ . Freshwater Biology , 46 : 795 – 805 .
- Smith , P. J. , McVeagh , S. M. and Collier , K. J. 2006 . ‘Genetic diversity and historical population structure in the New Zealand mayfly Acanthophlebia cruentata’ . Freshwater Biology , 51 : 12 – 24 .
- Snyder , C. D. and Hendricks , A. C. 1997 . ‘Genetic responses of Isonychia bicolor (Ephemeroptera:Isonychiidae) to chronic mercury pollution’ . Journal of the North American Benthological Society , 16 : 651 – 663 .
- Soldán , T. and Putz , M. 2000 . ‘Karyotypes of some Central European mayflies (Ephemeroptera) and their contribution ot phylogeny of the order’ . Acta Societatis Zoologicae Bohemoslovacae , 64 : 437 – 445 .
- Stahls , G. and Savolainen , E. 2008 . ‘MtDNA COI barcodes reveal cryptic diversity in the Baetis vernus group (Ephemeroptera, Baetidae)’ . Molecular Phylogenetics and Evolution , 46 : 82 – 87 .
- Sweeney , B. W. and Funk , D. H. 1991 . ‘Population genetics of the burrowing mayfly Dolania americana: geographic variation and the presence of a cryptic species’ . Aquatic Insects , 13 : 17 – 27 .
- Sweeney , B. W. , Funk , D. H. and Vannote , R. L. 1986 . ‘Population genetic structure of two mayflies (Ephemerella subvaria, Eurylophella verisimilis) in the Delaware River drainage basin’ . Journal of the North American Benthological Society , 4 : 253 – 263 .
- Sweeney , B. W. , Funk , D. H. and Vannote , R. L. 1987 . ‘Genetic variation in stream mayfly (Insecta: Ephemeroptera) populations of eastern North America’ . Annals of the Entomological Society of America , 80 : 600 – 612 .
- Sweeney , B. S. and Vannote , R. L. 1987 . ‘Geographic parthenogenesis in the stream mayfly Eurylophella funeralis in eastern North America’ . Holarctic Ecology , 10 : 52 – 59 .
- Takemon , Y. , Yamamoto , A. , Nakashima , M. , Tanida , K. , Kishi , M. and Kato , M. 2006 . ‘Isolation of sperm vesicles from adult male mayflies and other insects to prepare high molecular weight genomic DNA samples’ . Molecular Biology Reports , 33 : 65 – 70 .
- Thorpe , J. P. 1982 . ‘The molecular clock hypothesis: biochemical evolution, genetic differentiation and systematics’ . Annual Review of Ecology and Systematics , 13 : 139 – 168 .
- Tojo , K. and Matsukawa , K. 2003 . ‘A description of the second species of the family Dipteromimidae (Insecta, Ephemeroptera), and genetic relationship of two dipteromimid mayflies inferred from mitochondrial 16S rRNA gene sequences’ . Zoological Science , 20 : 1249 – 1259 .
- Vogler , A. P. and Monaghan , M. T. 2007 . ‘Recent advances in DNA taxonomy’ . Journal of Zoological Systematics and Evolutionary Research , 45 : 1 – 10 .
- Wang , T.-Q. and McCaffferty , W. P. 2004 . ‘Heptageniidae (Ephemeroptera) of the world. Part I: phylogenetic higher classification’ . Transactions of the American Entomological Society , 130 : 11 – 45 .
- Watanabe , K. and Omura , T. 2007 . ‘Relationship between reservoir size and genetic differentiation of the stream caddisfly Stenopsyche marmorata’ . Biological Conservation , 136 : 203 – 211 .
- Webb , J. M. , Sun , L. , McCafferty , W. P. and Ferris , V. R. 2007 . ‘A new species and new synonym in Heptagenia Walsh (Ephemeroptera: Heptageniidae: Heptageniinae) based on molecular and morphological evidence’ . Journal of Insect Science , 7 : 63
- Whitfield , J. B. and Kjer , K. M. 2008 . ‘Ancient rapid radiations of insects: challenges for phylogenetic analysis’ . Annual Review of Entomology , 53 : 449 – 472 .
- Wilcock , H. R. , Bruford , M. W. , Hildrew , A. G. and Nichols , R. A. 2005 . ‘Recruitment, kin and the spatial genetic structure of a caddisfly Plectrocnemia conspersa in a southern English stream’ . Freshwater Biology , 50 : 1499 – 1514 .
- Williams , H. C. , Ormerod , S. J. and Bruford , M. W. 2006 . ‘Molecular systematics and phylogeography of the cryptic species complex Baetis rhodani (Ephemeroptera, Baetidae)’ . Molecular Phylogenetics and Evolution , 40 : 370 – 382 .
- Wolf , E. 1960 . ‘Zur Karyologie der Eireifung und Forschung bei Cloeon dipterum L. (Bengtsson) (Ephemerida, Baetidae)’ . Biologisches Zentralblatt , 79 : 153 – 198 .
- Zhang , J. , Zhou , C. , Gai , Y. , Song , D. and Zhou , K. 2008 . ‘The complete mitochondrial genome of Parafronurus youi (Insecta: Ephemeroptera) and phylogenetic position of the Ephemeroptera’ . Gene , 424 : 18 – 24 .
- Zhou , C.-F. and Peters , J. G. 2003 . ‘The nymph of Siphluriscus chinensis and additional imaginal description: A living mayfly with Jurassic origins (Siphluriscidae new family: Ephemeroptera)’ . Florida Entomologist , 86 : 345 – 352 .
- Zickovich , J. M. and Bohonak , A. J. 2007 . ‘Dispersal ability and genetic structure in aquatic invertebrates: a comparative study in southern California streams and reservoirs’ . Freshwater Biology , 52 : 1982 – 1996 .
- Zloty , J. , Pritchard , G. and Krishnaraj , R. 1993 . ‘Larval insect identification by cellulose acetate gel electrophoresis and its application to life history evaluation and cohort analysis’ . Journal of the North American Benthological Society , 12 : 270 – 278 .
- Zurwerra , A. , Metzler , M. and Tomka , I. 1987 . ‘Biochemical systematics and evolution of the European Heptageniidae (Ephemeroptera)’ . Archiv für Hydrobiologie , 109 : 481 – 510 .
- Zurwerra , A. and Tomka , I. 1985 . ‘Electrogena gen. nov., eine neue Gattung der Heptageniidae (Ephemeroptera)’ . Entomologische Berichte Luzern , 13 : 99 – 104 .
- Zurwerra , A. , Tomka , I. and Lampel , G. ‘Application of the scanning electron microscope and the enzyme-gel-electrophoresis to solve taxonomical problems: the European species of Epeorus sensu Tshernova (1981)’ . Proceedings of the Fourth International Conference on Ephemeroptera . Edited by: Landa , T. , Soldán , V. and Tonner , M. pp. 213 – 218 . České Budějovice : Institute of Entomology, Czechoslovak Academy of Sciences .