Abstract
Background: As common contaminants of agricultural commodities, aflatoxins (AFs) are highly hazardous carcinogenic mycotoxins originating from very common fungi present in the environment. Their effect on bovine key immune-surveillance molecules is unclear.Objective: To determine the in vitro effects of naturally occurring level of AFs on toll-like receptor-4 (TLR4) and viability in bovine peripheral blood mononuclear cells (PBMCs) as a model system for immunotoxicity.Methods: Ten healthy dairy cows were selected as a source of PBMCs. The mRNA expression of TLR4 was quantitatively evaluated in PBMCs treated with a mix of 1.0, 0.5, 0.25 and 0.25 ng/ml of AFB1, AFB2, AFG1 and AFG2, respectively, and 10 ng/ml of the well-known TLR4 agonist lipopolysaccharide (LPS) for 0.5, 2, 5 and 18 hours. We also analyzed the viability of PBMCs exposed to AFs by means of the trypan blue exclusion method and the tetrazolium bromide assay.Results: Compared with control PBMCs, transcription of TLR4 in AFs-treated PBMCs was significantly up-regulated at post-exposure hours (PEH) 2 and 5, but was unchanged at PEH 0.5 and 18. Unsurprisingly, much higher up-regulation of TLR4 transcript was observed in LPS-treated PBMCs at PEH 0.5, 2, 5 and 18. Conversely, the viability of post-AFs and/or LPS-exposed PBMCs revealed no significant changes.Conclusion: Naturally occurring levels of AFs result in increased expression of TLR4 mRNA in bovine PBMCs.Clinical importance: The increased expression of TLR4 mRNA in bovine PBMCs by AFs also emphasizes the need to reduce admissible levels of AFs in feed for preventive measures.
1. Introduction
Aflatoxins (AFs) are highly hazardous feed-and-food-originated mycotoxins with potential redox and immune dysregulatory properties. They are produced by Aspergillus spp. present in the environment (Williams et al. Citation2004; Rawal et al. Citation2010; Chaytor et al. Citation2011), and their unanticipated consumption poses risks to animals and public health worldwide. Non-selective toxic effects of AFs including hepatotoxicity, cytotoxicity, genotoxicity and immunotoxicity in exposed animals and humans are well documented (Williams et al. Citation2004; Rawal et al. Citation2010; Chaytor et al. Citation2011; Mehrzad et al. Citation2011; Malvandi et al. Citation2013). Structurally, AFB2 and AFG2 are hydroxy derivatives of AFB1 and AFG1, respectively. Chemical structure of four common AFs reveals identical incorporation of dihydrofuran and tetrahydrofuran moieties coupled to a substituted coumarin. These four AFs, defined as total AFs, are found in common agricultural products, and harsh climate changes like gas emission, flood, drought and earthquake (in)directly ease the issue. In today's farm animals’ microenvironment, exposure to AFs by inhalation is very likely as well. Some other highly reactive and electrophilic metabolites of AFs like AFB1-epoxide, aflatoxicol, AFQ and AFM are also formed in vivo both in human and bovine, cumulatively causing more toxicity to the cells (Rawal et al. Citation2010; Mehrzad et al. Citation2011; Malvandi et al. Citation2013; Bahari et al. Citationin press). Hence, AFs in the environment can potentially pose considerable risk for bovine and human health.
The effects of clinically relevant levels of different AFs on the expression of TLR4 mRNA has been described in humans (Malvandi et al. Citation2013), but remains unknown in dairy cows. Toll-like receptors (TLRs) are responsible for sensing of a broad range of pathogen/damage-associated molecular patterns. TLR4 is the most studied and best characterized TLR in farm animals. It is highly expressed in leukocytes, and its activation, e.g., via AFs, leads to unnecessary production of many pro-inflammatory molecules (Liu & Wu Citation2010; Mehrzad et al. Citation2011; Malvandi et al. Citation2013; Bahari et al. Citationin press).
Food and feed is increasingly contaminated with AFs and food animals and humans usually consume a mix of different AFs. This happens more frequently in developing countries (Liu & Wu Citation2010; Shuaib et al. Citation2010). There is ample evidence for the role AFs play in both the innate and adaptive immune system in different animal species (Bernabucci et al. Citation2011; Chaytor et al. Citation2011; Mehrzad et al. Citation2011; Bruneau et al. Citation2012; Malvandi et al. Citation2013). What remains to be described is the effect of AFs on dairy cows. This study thus aimed to determine the effects of AFs on the expression of TLR4 mRNA in dairy cattle. We hypothesized that a clinically relevant mix of AFB1, AFB2, AFG1, AFG2 and lipopolysaccharide (LPS) will up-regulate TLR4 mRNA. Depending on the significance of the up-regulation, this will have consequences on the health of the cows. To verify our hypothesis on the interaction of mixed AFs and LPS with the bovine immune system, we determined the viability of PBMCs and measured the transcription of TLR4 by means of real-time quantitative PCR (qPCR).
2. Methods
2.1. Reagents, media, PBMCs isolation and in vitro experimental design
Fungal-derived AFs B1, B2, G1 and G2, and LPS of Escherichia coli 055:B5 were obtained from Sigma-Aldrich Chemie (Saint Louis, MO, USA); all of them were pure (>98%) and free of LPS and other chemicals except for LPS of E. coli 055:B5. Under sterile conditions, the AFs were first separately dissolved in 96% ethanol (0.1 mg/ml), and further diluted with sterile LPS-free Dulbecco's phosphate buffered saline (DPBS) (Mehrzad et al. Citation2011). Blood samples were aseptically collected into heparinized vacutainer tubes from 10 healthy dairy cows in mid-lactation (age 52.2 ± 1.7 (±standard error of means (SEM)) months). The PBMCs, which contain mainly a heterogeneous population of B and T lymphocytes and monocytes, were isolated accordingly (Mehrzad & Zhao Citation2008). Briefly, each blood sample was diluted 1:4 in DPBS (without Mg2+ and Ca2+) and then layered atop 15-ml Ficoll-Paque PLUS Lympholyte® (Zierikzee, the Netherlands) and centrifuged (1100 × g at 20 °C for 40 min). The layer of pure PBMCs was collected, washed (450 × g at 4 °C for 10 min) and the pellets were again washed (180 × g at 4 °C for 5 min) twice with RPMI-1640 containing 10% inactivated fetal calf serum (FCS), 1% penicillin/streptomycin, 1% kanamycin, and 1% gentamycin yielding >98% viable PBMCs with the trypan blue exclusion method (Mehrzad & Zhao Citation2008).
The isolated PBMCs from 10 cows were seeded in duplicate to 24-well tissue culture plates (2 ml/well RPMI medium with ∼6 × 106 PBMCs/well), containing 10% heat-inactivated FCS, 2 mM l-glutamine and 100 U/ml penicillin/streptomycin. The PBMCs incubated with mixed AFs (mix of 1.0, 0.5, 0.25 and 0.25 ng/ml of AFB1, AFB2, AFG1 and AFG2, respectively) and 10 ng/ml LPS (as positive control) for 0.5, 2, 5 and 18 hours (at 37 °C, 5% CO2, 95% humidity). The viability assay was performed on 20 μl of cell suspension from each well, and the PBMCs from each well were then centrifuged (350 × g at 4 °C for 5 min) for further molecular analysis.
2.2. RNA purification and cDNA synthesis
According to manufacturer's instruction, total RNA from treated and non-treated PBMCs was extracted and purified with TriPure isolation reagent (Roche Diagnostics, Indianapolis, IN, USA). The extracted RNA was then treated with DNase I kit (Fermentas, Vilnius, Lithuania) dissolved in nuclease-free ddH2O and the concentration and quality of the RNA was determined using nanodrop spectrophotometer and gel electrophoresis, respectively. About 0.5 μg of each RNA sample was reverse transcribed (at 65 °C for 5 min, 42 °C for 1 hour and 70 °C for 5 min) in a 20 μl final volume using a cDNA synthesis kit (Fermentas, Vilnius, Lithuania). The cDNA samples were then quantified with the nanodrop spectrophotometer.
2.3. Real-time quantitative reverse transcription-PCR assays
Before the quantitative reverse transcription (qRT)-PCR assays, gradient and conventional PCR was performed with the primers specific for bovine TLR4 and GAPDH, using an AccuPower® PCR PreMix (Seoul, Korea). Based on unconserved region and exon junction or separated exons strategies with Allele ID 7.5 (Premerbiosoft, CA, USA), the primers () were designed using CLC main workbench software (CLCbio Co, Aarhus, Denmark) to avoid mispairing during PCR assays. 5× HOT FIREPol® EvaGreen®qPCR Mix Plus (Solis BioDyne, Tartu, Estonia) with 6-Carboxyl-X-Rhodamine (ROX) was used for the qRT-PCR assays. Each reaction was 20 μl in final volume containing 30 ng of specific forward and reverse primers, 4 μl EvaGreen master mixes and 1 μl cDNA as templates. To optimize the qRT-PCR assays, different concentrations (0.5, 0.05, 0.005 and 0.0005 ng/μl) of cDNA were analyzed with those primers. The qRT-PCR conditions for both TLR4 and GAPDH were carried out, in duplicate, using light cycler system Rotter gen 6000 (Corbett Life Sciences, Mortlake, Australia)with cycling program including holding 15 min at 95 °C followed by cycling 45 times at 94, 55 and 72 °C and 20 sec for each temperature with melting curve analyses accompanied by agarose gel electrophoresis to ascertain the absence of non-specific PCR products.
Table1. Sequences, annealing temperature (Ta ) of designed primers, expected PCR fragment sizes (bp) and accession (Acc) numbers.
Normalization and analyses of qRT-PCR data were calculatedusing GenEX version 5 software (MultiD, Göteborg, Sweden) and Relative Expression Software Tool (REST; QIAGEN, Hilden, Germany). In each qRT-PCR reaction, the cycle number at which the fluorescence rose appropriately above background was determined as the crossing point (CP). Cycle threshold values were means of duplicates in each qRT-PCR run and 10 experimental repeats after interplate calibration. Optimization experiments were also performed to ensure that the efficiency of the target and the reference genes was approximately equal. The Pfaffl equation (Pfaffl Citation2001) was first used to calculate the relative gene expression ratio, i.e., the change in TLR4 expression divided by the change in GAPDH expression. A slope was determined from the exponential phase. The amplification efficiency (E) was calculated based on the slope, where E = 10[−1/slope]. The expression of TLR4 was calibrated to that of GAPDH using the formula: Relative fold change
2.4. Viability assay
The viability of mAFs-and/or LPS-exposed and control PBMCs was evaluated and confirmed using both manually trypan blue exclusion method (Mehrzad & Zhao Citation2008) by microscopic counting of 200 PBMCs with a hemocytometer as well as colorimetrically using absorbance at 570 nm with 3-(4,5-dimethylthiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Trypan blue exclusion and MTT methods were performed in 10 and 6 biological replicates, respectively.
2.5. Statistical analyses
After normalization of data calculated with GenEX version 5 software (MultiD, Göteborg, Sweden) and REST, the obtained results were expressed as means ± SEM. Statistical differences between control and mAFs/LPS-treated PBMCs were analyzed using student's t-test for difference between the two respective groups (i.e., at each time point the comparison was between mAFs/LPS-treated PBMCs and its respective control and/or non-treated PBMCs). A difference with p < 0.05 was considered statistically significant.
3. Results
The dynamics, detection and quantification of TLR4 gene expression in mAFs and/or LPS-exposed bovine PBMCs during different time points are depicted in . Dynamically, the analysis of the qPCR data revealed that clinically relevant levels of mAFs exposure to PBMCs resulted in a significant increase in expression of the TLR4 transcript, 2 hours (17-fold difference, p < 0.001) and 5 hours (9-fold difference, p < 0.001) after exposure (, left panel). The increased TLR4 transcription in LPS-treated PBMCs at postexposure hours (PEH) 0.5, 2, 5 and 18 was ∼7-fold, ∼29-fold, ∼26-fold and ∼15-fold higher than their untreated control counterparts, respectively, which was exceedingly higher than those of mAFs (, left panel). Constantly far more pronounced effects in LPS-treated PBMCs during all PEH was not surprising, chiefly because of the TLR4 agonistic property of LPS, and we used LPS as conveniently strong positive control for TLR4 transcriptomic analysis.
Figure1. The dynamics, detection and quantification of TLR4 gene expression (left panel) and viability (right panel) in mAFs and/or LPS-exposed bovine PBMCs during different time points. The differences between each group and its respective control (***p < 0.001). Neither mAFs nor LPS had negative effects on PBMCs viability. Mean (±SEM) values based on 10 dairy cows, and for the absorbance at 570 nm by MTT assay, see lower right panel (n = 6 biological repeats). RFC is relative fold change, i.e., relative to their non-treated counterparts.
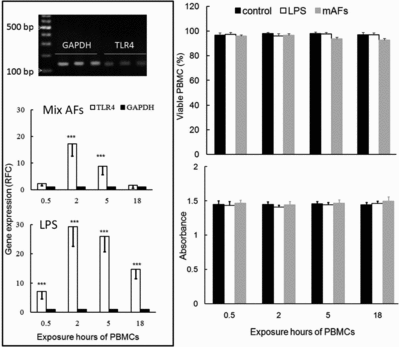
Next, to evaluate the viability of mAFs/LPS-exposed and non-exposed PBMCs in cell culture plates we performed the viability assay and observed no significant change on the viability and/or necrosis of mAFs-and/or LPS-treated PBMCs (, right panel).
4. Discussion
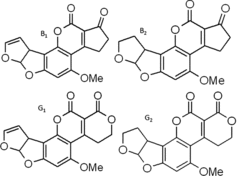
AFs are mainly classified into four types based on their structure: AFB1, AFB2, AFG1 and AFG2 (), which are all present on Aspergillus-infested crops. Although the exact proportion of AFB1, B2, G1 and G2 in Aspergillus-contaminated feed is unclear, these molds predominantly produce AFB1 and to a lesser extent the other three AFs in feed (Cervino et al. Citation2008; Li et al. Citation2009; Joubrane et al. Citation2011). We tested PBMCs with a dose of mAFs corresponding approximately to what may be found in mold-contaminated feed and hence could reflect naturally occurring levels in dairy cows. Indeed, when cows are fed feedstuffs contaminated at the upper limit of 10 μg/kg feed on a dry matter basis which is tolerated by the European Union feed legislation (2002) and considering the huge daily feed intake of high yielding dairy cows, the total AFs level in the blood stream of the exposed cow could reach ∼10 ng/ml. Moreover, far higher levels than the selected dose may be encountered by dairy cows in countries where mycotoxin monitoring programs are poor. Animals and humans could inevitably and chronically be exposed to levels of AFs that potentially cause significant toxic effects (Fink-Gremmels Citation1999; Martins et al. Citation2007; Pukkala et al. Citation2009; Shuaib et al. Citation2010) worldwide. In vivo AFs can be converted to even more reactive and electrophilic epoxides (Rawal et al. Citation2010; Mehrzad et al. Citation2011), like aflatoxicol, AFB1-epoxide, AFM1 and AFQ and AFs-related activation of the immune system in food animals is a serious issue (Bernabucci et al. Citation2011; Mehrzad et al. Citation2011; Mehrzad et al. Citationin press).
Figure2. Aflatoxins in feed are mainly classified into four types based on their structure: AFB1, AFB2, AFG1 and AFG2. OMe means methoxy.
TLR4 and relatively similarly CD14 and MyD88 are able to respond to potential oxidative conditions caused by AFs (Malvandi et al. Citation2013). Components of AFs metabolites, e.g., AFs-epoxide and aflatoxicol, which are produced in immune cells are highly oxidative and AFs themselves switch cells into the oxidative state (Bernabucci et al. Citation2011; Mehrzad et al. Citation2011; Malvandi et al. Citation2013; Bahari et al. Citationin press). Mechanistically, at this stage we do not know whether a direct result of the binding of AFs to the TLR4 or the oxidative state of the PBMCs induced up-regulated TLR4 mRNA expression. Basically, TLR4 recognizes its ligands with cooperation of cell surface proteins like CD14 and MD2. Consistent with findings by others (Ubagai et al. Citation2008; Mehrzad et al. Citation2011), our results clearly demonstrated that PBMCs immunologically sense AFs and in turn increase TLR4 mRNA. Bruneau et al. Citation(2012) failed to show up-regulation of TLR4 mRNA in murine macrophage cell line by AFs; this inconsistency might be due to a number of factors, such as cell type, animal species and AFs’ doses. It has been shown that AFs rapidly enter the leukocytes, and potentially bind to numerous xenobiotic sensing nuclear molecules like the pregnane X receptor, xenobiotic responsive enhancer module, etc., at the upstream region of cytochrome P450 isoforms (Ratajewski et al. Citation2011; Bahari et al. Citationin press), thereby potentiating pro-inflammatory–anti-inflammatory imbalance in PBMCs (Ratajewski et al. Citation2011; Malvandi et al. Citation2013). To our knowledge, it has not been established yet that LPS/AFs activate TLR4, and subsequently activate downstream signaling of TLR4 adaptor protein MyD88 leading to (in)dependent activation of NF-κB, eventually causing secretion of pro-inflammatory cytokines. Despite of those pathways we hardly observed necrosis in mAFs-exposed PBMCs, which needs further clarification. Measuring the protein content of TLR4 or the concentration of proinflammatory cytokines following TLR4 activation or transcriptomics analyses of some TLR4 signaling pathway mediators such as MyD88, NF-κB, etc. in PBMCs seems valuable.
Despite the prolonged over-expression of TLR4 transcript, and subsequently more oxidative stress as reported by Mehrzad et al. Citation2011, the viability of mAFs/LPS-exposed PBMCs underwent little change. Similarly, AFs had little effect on necrosis and apoptosis of bovine neutrophils (Mehrzad et al. Citation2011). This study provides essential basic knowledge on the role that TLR4 potentially plays in the recognition of AFs. We expect that AFs could signal through TLR4. Nevertheless future research should be directed towards elucidation of the disturbing effects AFs might have on innate and adaptive immunity. The concept of AFs-mediated redox imbalance in situ (Ubagai et al. Citation2008; Mehrzad et al. Citation2011) should not be ignored either.
Briefly, the presented results show that even environmentally relevant levels of AFs result in the increased expression of TLR4 mRNA in bovine PBMCs.
Acknowledgements
This work was financially supported by a grant from Ferdowsi University of Mashhad, Iran. The authors thank Mr A. Bahari for technical support.
References
- Bahari A, Mehrzad J, Mahmoudi M, Bbassami MR, Dehghani H. in press. Cytochrome P450 isoforms are differently up-regulated in aflatoxin B1-exposed human lymphocytes and monocytes. Immunopharmacol Immunotoxicol. doi:10.3109/08923973.2013.850506
- Bernabucci U, Colavecchia L, Danieli PP, Basirico L, Lacetera N, Nardone A, Ronchi B. 2011. Aflatoxin B1 and fumonisin B1 affect the oxidative status of bovine peripheral blood mononuclear cells. Toxicol In Vitro. 25:684–691.
- Bruneau JC, Stack E, O’Kennedy R, Loscher CE. 2012. Aflatoxins B(1), B(2) and G(1) modulate cytokine secretion and cell surface marker expression in J774A.1 murine macrophages. Toxicol In Vitro. 26:686–693.
- Cervino C, Asam S, Knopp D, Rychlik M, Niessner R. 2008. Useof isotope-labeled aflatoxins for LC-MS/MS stable isotope dilution analysis of foods. J Agric Food Chem. 56:1873–879.
- Chaytor AC, See MT, Hansen JA, de Souza AL, Middleton TF, Kim SW. 2011. Effects of chronic exposure of diets with reduced concentrations of aflatoxin and deoxynivalenol on growth and immune status of pigs. J Anim Sci. 89:124–135.
- Fink-Gremmels J. 1999. Mycotoxins: their implications for human and animal health. Vet Q. 21:115–150.
- Joubrane K, El Khoury A, Lteif R, Rizk T, Kallassy M, Hilan C, Maroun R. 2011. Occurrence of aflatoxin B1 and ochratoxin A in Lebanese cultivated wheat. Mycotoxin Res. 27:249–57.
- Li FQ, Li YW, Wang YR, Luo XY. 2009. Natural occurrence of aflatoxins in Chinese peanut butter and sesame paste. J Agric Food Chem. 57:3519–3524.
- Liu Y, Wu F. 2010. Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ Health Perspect. 118:818–824.
- Malvandi AM, Mehrzad J, Saleh-moghaddam M. 2013. Biologically relevant doses of mixed aflatoxins B and G up-regulate MyD88, TLR2, TLR4 and CD14 transcripts in human PBMCs. Immunopharmacol Immunotoxicol. 35:528–532.
- Martins HM, Mendes Guerra MM, d’Almeida Bernardo FM. 2007. Occurrence of aflatoxin B1 in dairy cow feed over 10 years in Portugal (1995–2004). Rev Iberoam Micol. 24:69–671.
- Mehrzad J, Devriendt B, Baert K, Cox E. in press. Aflatoxin B1 interferes with the antigen-presenting capacity of porcine dendritic cells. Toxicol In Vitro.
- Mehrzad J, Klein G, Kamphues J, Wolf P, Grabowski N, Schuberth HJ. 2011. In vitro effects of very low levels of aflatoxin B1 on free radicals production and bactericidal activity of bovine blood neutrophils. Vet Immunol Immunopathol. 141:16–25.
- Mehrzad J, Zhao X. 2008. T-lymphocyte proliferative capacity and CD4+/CD8+ ratio in primiparous and pluriparous lactating cows. J Dairy Res. 75:457–465.
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
- Pukkala E, Martinsen JI, Lynge E, Gunnarsdottir HK, Sparén P, Tryggvadottir L, Weiderpass E, Kjaerheim K. 2009. Occupation and cancer-follow-up of 15million people in five Nordic countries. Acta Oncol. 48:646–690.
- Ratajewski M, Walczak-Drzewiecka A, Salkowska A, Dastych J. 2011. Aflatoxins upregulate CYP3A4 mRNA expression in a process that involves the PXR transcription factor. Toxicol Lett. 205:146–153.
- Rawal S, Kim JE, Coulombe R Jr. 2010. Aflatoxin B1 in poultry: toxicology, metabolism and prevention. Res Vet Sci. 89:325–331.
- Shuaib FM, Ehiri J, Abdullahi A, Williams JH, Jolly PE. 2010. Reproductive health effects of aflatoxins: a review of the literature. Reprod Toxicol. 29:262–270.
- Ubagai T, Tansho S, Ito T, Ono Y. 2008. Influences of aflatoxin B1 on reactive oxygen species generation and chemotaxis of human polymorphonuclear leukocytes. Toxicol In Vitro. 22:1115–1120.
- Williams JH, Phillips TD, Jolly PE, Stiles JL, Jolly CM Aggarwal D. 2004. Human aflatoxicosis in developing countries: A review of toxicology, exposure, potential health consequences, and interventions. Am J Clin Nutr. 80:1106–1122.