ABSTRACT
Background: Parapoxviruses are zoonotic viruses that infect cattle, goats and sheep; there have also been reports of infections in camels, domestic cats and seals.
Objective: The objective of this report was to describe a case of vesicular disease caused by pseudocowpox virus (PCPV) in water buffalo (Bubalus bubalis) in Brazil.
Animals: Sixty buffalo less than 6 months old exhibited ulcers and widespread peeling of the tongue epithelium. There were no cases of vesicular disease in pigs or horses on the same property.
Methods: Samples were analysed by PCR and sequencing. Phylogenetic analysis in MEGA 7.01 was reconstructed using major envelope protein (B2L) by the Tamura three-parameter nucleotide substitution model and the maximum likelihood and neighbor joining models, both with 1000 bootstrap replicates. The genetic distance between the groups was analysed in MEGA using the maximum composite likelihood model. The rate variation among sites was modeled using gamma distribution.
Results: The presence of PCPV in the buffalo herd could be demonstrated in epithelium and serum. The minimum genetic distance between the isolated PCPV strain (262-2016) and orf virus and bovine papular stomatitis virus was 6.7% and 18.4%, respectively. The maximum genetic distance calculated was 4.6% when compared with a PCPV detected in a camel.
Conclusions/Clinical Importance: The peculiar position of the isolated strain in the phylogenetic trees does not necessarily indicate a different kind of PCPV that infects buffalo. More samples from cattle and buffalo in Brazil must be sequenced and compared to verify if PCPV from buffalo are genetically different from samples derived from cattle.
1. Introduction
Parapoxvirus (PPV) is a genus belonging to the Poxviridae family that comprises four virus species currently recognized: pseudocowpox virus (PCPV), bovine papular stomatitis virus (BPSV), orf virus (ORFV) and PPV of red deer in New Zealand (PVNZ) (Airas et al. Citation2016). These viruses are gaining importance because of their zoonotic potential and because of financial and economic losses connected with disease outbreaks in livestock.
These viruses infect cattle, goats and sheep, but there have been reports of infections in camels and seals too (Essbauer et al. Citation2010). All known PPVs can infect humans after direct or indirect contact with infected animals, and result of the infection is local skin lesions (Friederichs et al. Citation2014). Virus replication occurs in epidermal keratocytes, producing pustular lesions in regions such as mouth, tongue, lips or teats. Typically, each case resolves in 4–6 weeks, but complications may occur due to secondary infections by bacteria (Inoshima et al. Citation2000).
PPVs are double-stranded DNA viruses with a genome size varying from 130 to 150 kbp with a guanine-cytosine content of roughly 64%. Isolates were defined only by host origin, but an increasing number of studies are now using molecular data such as whole-genome sequencing to define new species (Mercer et al. Citation2006). PPVs have 88 genes in the central region of their genome, and most of the unique and pathogenic genes are located in the terminal regions (Mercer et al. Citation2006). The preferred gene for sequence analysis is B2L (ORF011), an ortholog of the vaccinia virus gene F13L that encodes the major envelope antigen p37K (Sullivan et al. Citation1994).
PPVs are also important in the differential diagnosis of vesicular diseases such as foot and mouth disease (FMD) and vesicular stomatitis virus. Farms with a suspicion of these diseases may be subject to strict rules regarding the transit of animals and products derived from animals, which may cause economic problems. A previous study described multiple viral infection in cases of vesicular diseases in Brazil with one sample collected that was positive for PCPV (Laguardia-Nascimento et al. Citation2016). Buffalo are susceptible to poxvirus, but typically by orthopoxvirus (OPV) such as vaccinia virus (VACV) and Buffalopox virus (BPXV) (Franco-Luiz et al. Citation2016).
The objective of this work is to describe a case of PCPV infection in water buffalo (Bubalus bubalis) in São Paulo, Brazil.
2. Material and methods
2.1. Establishment
The Official Veterinary Service responded to a case of suspected vesicular disease on a dairy farm with 122 water buffalo in the city of Barra do Turvo in the state of São Paulo, Brazil.
Information about the farm was obtained from questionnaires. The buffalo were vaccinated for FMD following the recommendations of the Brazilian legislation. In terms of contact with neighboring farms, the site was used for tourism activities and it did not share equipment with other establishments. There was inflow of vehicles for milk collection, and people visited who had been exposed to susceptible animals at other establishments. There was no change in the supply or management history of the farm, but there was a historical record of ingestion of toxic plants leading to clinical symptoms. Nobody working on the farm had visited other countries within the last 30 days.
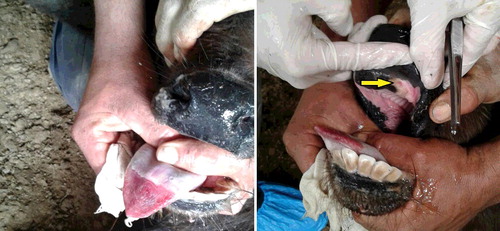
Sixty buffalo less than 6 months old exhibited apathy, intense drooling, ulcers and widespread peeling of the tongue epithelium (). One calf died. There was no case of vesicular disease in pigs, horses or adult buffalo on the same property. The nearest cattle herd was located 15 km away. It was reported that vehicles used for milk transportation drove between herds. The farm was closed by the Official Veterinary Service (i.e. animals and related products did not leave or arrive).
2.2. Molecular diagnostics
Four epithelium tissues collected from four animals were sent to a laboratory to investigate vesicular disease. The samples were examined in an OIE level 4 biosafety laboratory due the possibility of infection by FMD virus (FMDV). RNA was extracted using TRIzol (Invitrogen, Waltham, MA, USA) and DNA was extracted using DNAeasy Blood and Tissue (Qiagen, Hilden, Germany), as recommended by the manufacturer. Acid nucleic extraction was evaluated by detecting the gene or messenger RNA of beta-actin. Molecular tests were conducted to detect bovine viral diarrhea virus, vesicular stomatitis virus (VSV) (protocol used for detecting the subtypes Alagoas, Indiana, New Jersey and Cocal), ovine herpesvirus 2, blue tongue virus, OPV, PPV and bovine herpesvirus 1. All of the protocols were conducted as described previously in the literature (Laguardia-Nascimento et al. Citation2016).
2.3. Virus isolation and serological tests
The same tissue samples were subjected to virus isolation in MDBK and BHK21 (Abrahão et al. Citation2015). Serum was collected from 58 animals for the detection of antibodies against FMDV using ELISA and for the detection of antibodies against all four VSV serotypes using serum neutralization, as described previously in the literature (Laguardia-Nascimento et al. Citation2016).
2.4. Molecular phylogeny
Positive samples were sent to sequencing to confirm the species of the PPV detected and submitted to GenBank (accession number KX980495). Phylogenetic analysis in MEGA 7.01 (Kumar et al. Citation2016) was reconstructed using major envelope protein (B2L) using the Tamura three-parameter nucleotide substitution model and the maximum likelihood (ML) and neighbor joining (NJ) models, both with 1000 bootstrap replicates. The genetic distance between the groups was analysed in MEGA using the maximum composite likelihood model. The rate variation among sites was modeled using a gamma distribution. Sequences deposited in GenBank are listed in
Table 1. Sequences used in this work to construct phylogenetic trees.
3. Results
3.1. Diagnostics
All of the serological and virus isolation tests were negative. The molecular tests were negative for all viruses except for PCPV (serum was also subjected to PCR and PCPV were detected in two samples).
3.2. Molecular phylogeny
The sequence from the affected buffalo (262-2016) clustered with other PCPV detected in cattle, camel and reindeer but in an isolated clade. The minimum genetic distance between 262-2016 and other PCPV from Brazil was 2.4%, and the minimum genetic distance between 262-2016 and ORFV and BPSV was 6.7% and 18.4%, respectively. The maximum genetic distance calculated was 4.6% when compared with a PCPV detected in a camel. Sequences from the same countries or from similar regions in Brazil clustered together more tightly, but this result appears to be biased since it derives from samples collected from the same outbreaks. There are samples from Cameroon and Turkey in the same clade as Germany with genetic distances as low as 0.2%.
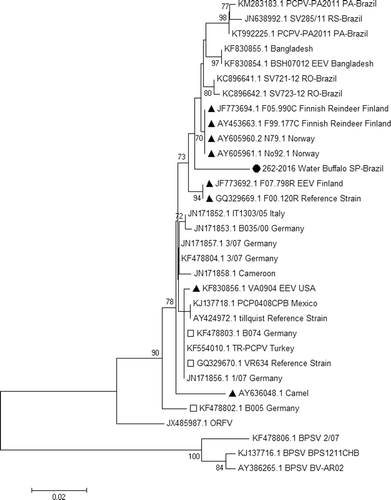
The constructed trees had similar topologies but with significant changes in the 262-2016 position. This sequence clustered with other PCPV detected in cattle in Rondonia state in Brazil when the ML model was used (). However, this sequence formed an isolated clade in the NJ tree (). In both trees, 262-2016 was still distant from the other samples. The genetic distance was not strongly correlated with geographic distance in the phylogenetic tree. There are samples from distant countries such as Cameroon, Germany and Mexico that clustered in the same clade. Sequences from humans grouped with other samples from cattle.
Figure 2. Phylogenetic analysis in MEGA 7.01 was reconstructed using major envelope protein (B2L) by the Tamura three-parameter nucleotide substitution model and the maximum likelihood model with 1000 bootstrap replicates. Strain 262-2016 isolated from the affected Water buffalo (Bubalus bubalis) in this study clustered with other samples from Brazil but with higher genetic distance than others in the same cluster.
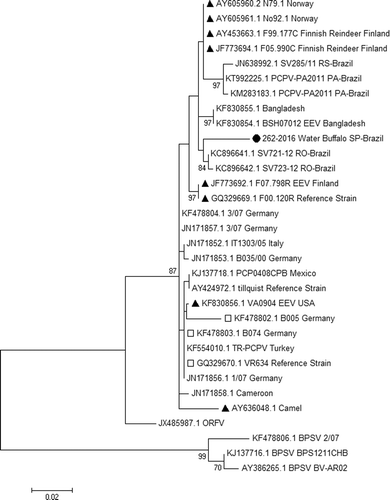
Figure 3. Phylogenetic analysis in MEGA 7.01 was reconstructed using major envelope protein (B2L) using the Tamura three-parameter nucleotide substitution model and neighbor joining model with 1000 bootstrap replicates. Strain 262-2016 isolated from the affected water buffalo (Bubalus bubalis) in this study was positioned in an isolated clade, distant from other samples from Brazil.
4. Discussion
Water buffalo breeding is an important economic alternative to cattle breeding in Brazil. These animals are better adapted to wet tropical and subtropical environments than cattle. They are an important resource of proteins for human consumption in several regions of the world such as South America, Africa and Asia. The size of the buffalo population increases in Brazil at an annual rate of 12%, and these increases occur largely in damp areas not suitable for cattle (Vasconcelos et al. Citation2001). There are approximately 2.8 million buffalo distributed across all Brazilian states, and the global population of this species is estimated to be more than 174 million. Buffalo have exhibited more population growth than cattle in recent years in Brazil (Malhado et al. Citation2013).
Viruses that infect buffalo may also affect cattle. Bovine herpesvirus 1, for example, is wide spread in cattle and may cross the species barrier and infect buffalo (Caruso et al. Citation2016). Bovine viral diarrhea virus is another important pathogen that primarily infects ruminants, and it leads to several clinical problems such as abortion. Bovine viral diarrhea virus is also widespread in cattle and may affect buffalo (Craig et al. Citation2015).
Brazilian veterinary services maintain a constant vigilance in cases of vesicular disease in various species. There have been no reported outbreaks of FMDV since 2005, but other viruses such as OPV, VSV and PPV are still present in cattle (Cargnelutti et al. Citation2014; Abrahão et al. Citation2015), swine (Medaglia et al. Citation2011) and buffalo (Franco-Luiz et al. Citation2016). Collected samples are subjected to thorough investigations to better understand the causes of clinical signs.
PCPV appears to be a new threat to cattle and buffalo sharing space in Brazil. PCPV infections in cattle are spreading to buffalo. The first case was reported in Amazon (North region), in an area in which mixed populations of cattle and buffalo were common. This study describes another case in São Paulo (Southeast region) with no epidemiological link to the previous report. This investigation focuses on a dairy farm with only buffalo and no bovines. The presence of buffalo and cattle on the same farms in Brazil raises the possibility of cross-species transmission of PCPV; it has been shown that these viruses can cross species barriers (Klein & Tryland Citation2005).
The peculiar position of 262-2016 in the phylogenetic trees does not necessarily indicate a different kind of PCPV infecting buffalo. More samples from cattle and buffalo from Brazil must be sequenced and compared to verify if PCPV from buffalo are genetically different from samples derived from cattle. PCPVs from reindeer have genetic differences compared with samples from cattle, but they are still grouped in PCPV clades; there does not appear to be a strain specialized in that species (Hautaniemi et al. Citation2011). PPVs detected in camel also group in different clades, but they are not monophyletic, which also indicates that their genetic variation is not related to specialization to a single host (Khalafalla et al. Citation2015). These analyzes should be carried out carefully because, as seen in this work, the choice of model can change the position of the samples.
PPVs are now part of the molecular tests for differential diagnostics of FMD in Brazil. The presence of these viruses in different species has been detected in samples sent to official laboratories for tests of vesicular diseases (Laguardia-Nascimento et al. Citation2016). Veterinarians typically collect only the epithelium from lesions for molecular tests. However, in this work we also found the virus in serum. The presence of viral DNA in serum also occurs with other poxviruses such as the VACV (Putkuri et al. Citation2009). Molecular tests may be an important tool to detect animals without symptoms since such animals also transmit this zoonotic agent (Yaegashi et al. Citation2016).
5. Conclusions
Our diagnostic and genetic analysis supports the involvement of PCPV in buffalo suffering from vesicular disease. Veterinarians should be aware of the possibility of vesicular disease in buffalo caused by PCPV, and specific tests should be included in differential diagnoses of vesicular diseases in this species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Abrahão JS, Campos RK, Trindade G de S, Guimarães da Fonseca F, Ferreira PC, Kroon EG. 2015. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg Infect Dis. 21:695–698.
- Airas N, Hautaniemi M, Syrjä P, Knuuttila A, Putkuri N, Coulter L, McInnes CJ, Vapalahti O, Huovilainen A, Kinnunen PM. 2016. Infection with possible novel parapoxvirus in horse, Finland, 2013. Emerg Infect Dis. 2:1242–1245.
- Cargnelutti JF, Flores MM, Teixeira FR, Weiblen R, Flores EF. 2012. An outbreak of pseudocowpox in fattening calves in southern Brazil. J Vet Diagn Invest. 24:437–441.
- Cargnelutti JF, Olinda RG, Maia LA, de Aguiar GM, Neto EG, Simões SV, de Lima TG, Dantas AF, Weiblen R, Flores EF, Riet-Correa F. 2014. Outbreaks of Vesicular stomatitis Alagoas virus in horses and cattle in northeastern Brazil. J Vet Diagn Invest. 26:788–794.
- Caruso C, Prato R, Ingravalle F, Vecchio D, Sciarra A, Ternavasio M, Ceccarelli L, Martucciello A, Galiero G, De Carlo E, Masoero L. 2016. Prevalence of antibodies against Bubaline herpesvirus (BuHV-1) among Mediterranean water buffalo (Bubalus bubalis) with implications in buffalo trade. Vet Q. 18:1–5.
- Craig MI, König Guido A, Benitez Daniel F, Draghi María G. 2015. Molecular analyses detect natural coinfection of water buffaloes (Bubalus bubalis) with bovine viral diarrhea viruses (BVDV) in serologically negative animals. Rev Argent Microbiol. 47:148–151.
- Dal Pozzo F, Martinelle L, Gallina L, Mast J, Sarradin P, Thiry E, Scagliarini A, Büttner M, Saegerman C. 2011. Original findings associated with two cases of bovine papular stomatitis. J Clin Microbiol. 49:4397–4400.
- Delhon G, Tulman ER, Afonso CL, Lu Z, de la Concha-Bermejillo A, Lehmkuhl HD, Piccone ME, Kutish GF, Rock DL. 2004. Genomes of the parapoxviruses ORF virus and bovine papular stomatitis virus. J Virol. 78:168–177.
- Essbauer S, Pfeffer M, Meyer H. 2010. Zoonotic poxviruses. Vet Microbiol. 140:229–236.
- Franco-Luiz AP, Fagundes Pereira A, de Oliveira CH, Barbosa JD, Oliveira DB, Bonjardim CA, Ferreira PC, de Souza Trindade G, Abrahão JS, Kroon EG. 2016. The detection of Vaccinia virus confirms the high circulation of Orthopoxvirus in buffaloes living in geographical isolation, Marajó Island, Brazilian Amazon. Comp Immunol Microbiol Infect Dis. 46:16–19.
- Friederichs S, Krebs S, Blum H, Wolf E, Lang H, von Buttlar H, Büttner M. 2014. Comparative and retrospective molecular analysis of Parapoxvirus (PPV) isolates. Virus Res. 181:11–21.
- Guo J, Rasmussen J, Wünschmann A, de La Concha-Bermejillo A. 2004. Genetic characterization of orf viruses isolated from various ruminant species of a zoo. Vet Microbiol. 99:81–92.
- Hautaniemi M, Ueda N, Tuimala J, Mercer AA, Lahdenperä J, McInnes CJ. 2010. The genome of pseudocowpoxvirus: comparison of a reindeer isolate and a reference strain. J Gen Virol. 91:1560–1576.
- Hautaniemi M, Vaccari F, Scagliarini A, Laaksonen S, Huovilainen A, McInnes CJ. 2011. Analysis of deletion within the reindeer pseudocowpoxvirus genome. Virus Res. 160:326–332.
- Inoshima Y, Morooka A, Sentsui H. 2000. Detection and diagnosis of parapoxvirus by the polymerase chain reaction. J Virol Methods. 84:201–208.
- Khalafalla AI, El-Sabagh IM, Al-Busada KA, Al-Mubarak AI, Ali YH. 2015. Phylogenetic analysis of eight sudanese camel contagious ecthyma viruses based on B2L gene sequence. Virol J. 12;124–132.
- Klein J, Tryland M. 2005. Characterisation of parapoxviruses isolated from Norwegian semi-domesticated reindeer (Rangifer tarandus tarandus). Virol J. 2:79–288.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Laguardia-Nascimento M, Sales ÉB, Gasparini MR, de Souza NM, da Silva JA, Souza GG, Carani FR, Dos Santos AF, Rivetti Júnior AV, Camargos MF, Fonseca Júnior AA. 2016. Detection of multiple viral infections in cattle and buffalo with suspected vesicular disease in Brazil. J Vet Diagn Invest. 28:377–381.
- Lederman E, Khan SU, Luby S, Zhao H, Braden Z, Gao J, Karem K, Damon I, Reynolds M, Li Y. 2014. Zoonotic parapoxviruses detected in symptomatic cattle in Bangladesh. BMC Res Notes. 19;7:816.
- Malhado CHM, Malhado ACM, Ramos AA, Carneiro PLS, Souza JC, Pala A. 2013. Genetic parameters for milk yield, lactation length and calving intervals of Murrah buffaloes from Brazil. Rev Bras Zoot. 42:565–569.
- Medaglia ML, Pereira Ade C, Freitas TR, Damaso CR. 2011. Swinepox virus outbreak, Brazil, 2011. Emerg Infect Dis. 17:1976–1978.
- Mercer AA, Ueda N, Friederichs SM, Hofmann K, Fraser KM, Bateman T, Fleming SB. 2006. Comparative analysis of genome sequences of three isolates of Orf virus reveals unexpected sequence variation. Virus Res. 116:146–158.
- Oğuzoğlu TÇ, Koç BT, Kirdeci A, Tan MT. 2014. Evidence of zoonotic pseudocowpox virus infection from a cattle in Turkey. Virusdisease. 25:381–384.
- Putkuri N, Piiparinen H, Vaheri A, Vapalahti O. 2009. Detection of human orthopoxvirus infections and differentiation of smallpox virus with real-time PCR. J Med Virol. 81:146–152.
- Schmidt C, Cargnelutti JF, Brum MC, Traesel CK, Weiblen R, Flores EF. 2013. Partial sequence analysis of B2L gene of Brazilian orf viruses from sheep and goats. Vet Microbiol. 162:245–253.
- Sullivan JT, Mercer AA, Fleming SB, Robinson AJ. 1994. Identification and characterization of an orf virus homologue of the vaccinia virus gene encoding the major envelope antigen p37K. Virology. 202:968–973.
- Tikkanen MK, McInnes CJ, Mercer AA, Büttner M, Tuimala J, Hirvelä-Koski V, Neuvonen E, Huovilainen A. 2004. Recent isolates of parapoxvirus of Finnish reindeer (Rangifer tarandus tarandus) are closely related to bovine pseudocowpox virus. J Gen Virol. 85:1413-–1418.
- Vasconcellos SA, Oliveira JCF, Morais ZM, Baruselli PS, Amaral R, Pinheiro SR. 2001. Isolation of Leptospira santarosai, serovar guaricura from buffaloes (Bubalus bubalis) in Vale do Ribeira, São Paulo, Brazil Braz J Microbiol. 32:298–300.
- Yaegashi G, Fukunari K, Oyama T, Murakami RK, Inoshima Y. 2016. Detection and quantification of parapoxvirus DNA by use of a quantitative real-time polymerase chain reaction assay in calves without clinical signs of parapoxvirus infection. Am J Vet Res. 77:383–387.