Abstract
Swine coronaviruses (SCoVs) are one of the most devastating pathogens affecting the livelihoods of farmers and swine industry across the world. These include transmissible gastroenteritis virus (TGEV), porcine epidemic diarrhea virus (PEDV), porcine respiratory coronavirus (PRCV), porcine hemagglutinating encephalomyelitis virus (PHEV), swine acute diarrhea syndrome coronavirus (SADS-CoV), and porcine delta coronavirus (PDCoV). Coronaviruses infect a wide variety of animal species and humans because these are having single stranded-RNA that accounts for high mutation rates and thus could break the species barrier. The gastrointestinal, cardiovascular, and nervous systems are the primary organ systems affected by SCoVs. Infection is very common in piglets compared to adult swine causing high mortality in the former. Bat is implicated to be the origin of all CoVs affecting animals and humans. Since pig is the only domestic animal in which CoVs cause a wide range of diseases; new coronaviruses with high zoonotic potential could likely emerge in the future as observed in the past. The recently emerged severe acute respiratory syndrome coronavirus virus-2 (SARS-CoV-2), causing COVID-19 pandemic in humans, has been implicated to have animal origin, also reported from few animal species, though its zoonotic concerns are still under investigation. This review discusses SCoVs and their epidemiology, virology, evolution, pathology, wildlife reservoirs, interspecies transmission, spill-over events and highlighting their emerging threats to swine population. The role of pigs amid ongoing SARS-CoV-2 pandemic will also be discussed. A thorough investigation should be conducted to rule out zoonotic potential of SCoVs and to design appropriate strategies for their prevention and control.
1. Introduction
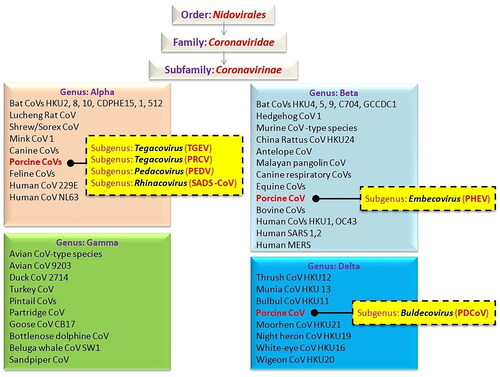
The transmission of zoonotic diseases has become the world’s most serious threat to public health. As pathogens of importance on a global level, zoonotic CoVs have a higher tendency for cross-species transmission to humans and other important domesticated species (Edwards et al. Citation2020). Coronaviruses (CoVs) cause a variety of diseases in both animals and humans, and their ability to cause dreadful diseases in livestock and companion animals prompted extensive research in the twentieth century. Coronaviruses are a wide group of mammalian and avian pathogens identified in 1968 (Almeida et al. Citation1968; Alluwaimi et al. Citation2020). They belong to the order Nidovirales, Coronaviridae family, and Coronavirinae subfamily (Alluwaimi et al. Citation2020). Since CoVs infect a broad range of habitats, several species of animals carry these pathogens as well as affects humans; causing respiratory, enteric, or systemic diseases in a variety of mammalian hosts that vary in clinical severity from subclinical, mild, moderate to fatal (Cui et al. Citation2019; Andersen et al. Citation2020; Dhama et al. Citation2020; Ghosh and Malik Citation2020; Haake et al. Citation2020; Colina et al. Citation2021; Zhang et al. Citation2021a). Coronaviruses have been reported to cause infection in diverse species of animals including cattle, swine, horses, cats, dogs, camels, rabbits, rodents, bats, palm civets, ferrets, mink, snake, avian/poultry species as well as wildlife (Kahn and McIntosh Citation2005; Dhama et al. Citation2014a, Citation2014b; Fehr and Perlman Citation2015; Miłek and Blicharz-Domańska Citation2018; Haake et al. Citation2020; Malik et al. Citation2020; Colina et al. Citation2021; Zhang et al. Citation2021a). During the past two decades, three deadly and most pathogenic CoVs have affected humans namely, severe acute respiratory syndrome coronavirus (SARS-CoV) (2003), Middle East respiratory syndrome coronavirus (MERS-CoV) (2012), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (2019), the last one as the cause of ongoing coronavirus disease 2019 (COVID-19) pandemic (Ksiazek et al. Citation2003; Berger et al. Citation2004; Dhama et al. Citation2020; Rabaan et al. Citation2020, Citation2021; Shaw et al. Citation2020).
Swine CoVs (SCoVs) are the major enteric and respiratory pathogens in swine. Nevertheless, cardiovascular, gastrointestinal, as well as peripheral and/or central nervous system illnesses are also noticeable in the vast majority of instances. Four CoV genera have been identified based on genomic criteria: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Chen et al. Citation2020). Six swine CoVs have been reported among them: (1) the transmissible gastroenteritis virus (TGEV), (subgenus Tegacovirus) which was first identified in 1946; (2) the porcine hemagglutinating encephalomyelitis virus (PHEV) (subgenus Embecovirus) identified in 1957; (3) the porcine epidemic diarrhea virus (PEDV) (subgenus Pedacovirus), found in 1977; (4) the porcine respiratory coronavirus (PRCV), a variant of TGEV discovered in 1984; (5) the porcine delta coronavirus (PDCoV) (subgenus Buldecovirus) reported in 2012; and (6) swine acute diarrhea syndrome coronavirus (SADS-CoV) (subgenus Rhinacovirus), discovered in 2017 (Decaro et al. Citation2020). Detailed and systematic classification of CoVs is depicted in . Four of the Swine coronavirus, i.e. TGEV, PRCV, PEDV, SADS-CoV belong to genus Alphacoronavirus, PHEV to genus Betacoronavirus while PDCoV to genus Deltacoronavirus. Concomitant infection with TGEV, PDCoV, PEDV and SADS-CoV are common in infected pig farms resulting in acute gastroenteritis (Luo et al. Citation2020). However, PRCV can also cause pneumonia (Boley et al. Citation2020). The following factors contribute to increased susceptibility of neonatal pigs to CoV infection namely: the immune system of a young pig is not completely developed; the stomach pH of younger pigs is less acidic than that of older pigs; the regeneration of enterocytes that line the intestinal villi through progenitor cells is slower in younger pigs; piglets are more vulnerable to fluid and electrolyte imbalances due to mal-digestion and intense malabsorption (Doyle and Hutchings Citation1946; Turlewicz-Podbielska and Pomorska-Mól Citation2021). Porcine epidemic diarrhea virus is considered to be more pathogenic than PDCoV, and the mortality rates of PEDV and PDCoV in newborn piglets are about 80–100% and 40%, respectively (Jung et al. Citation2016; Niederwerder and Hesse Citation2018). In China, SADS-CoV mortality was high (90–100%) in piglets younger than 5 days old, but it was reduced to 5% in piglets older than 8 days (Zhou et al. Citation2018; Xu et al. Citation2019).
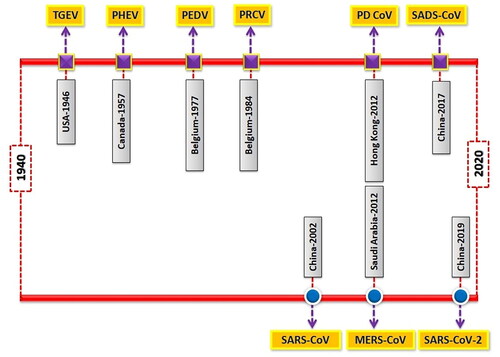
Outbreaks of SCoVs are more common in winter and spring seasons. Piglets are more susceptible to infection. The disease is characterized by severe clinical symptoms such as diarrhea, vomiting, anorexia, dehydration, metabolic acidosis, hyperkalemia and further it may lead to death. Villus destruction and multifocal necrosis of intestinal epithelium are the most noticeable pathological features of SCoV infection (Luo et al. Citation2017). Coronaviruses occur as quasispecies with a high degree of genetic recombination and mutation (Forni et al. Citation2017). This promotes the introduction of newer strains with changes in cell tropism and host specificity. Virus spreading from wild animal sources is a significant threat to human and animal health (Olival et al. Citation2017). The majority of SCoVs are transmitted by fecal-oral route, whereas PRCV is transmitted primarily through inhalation (Saade et al. Citation2020). Bats have been identified as the primary reservoirs for emerging coronaviruses (Zhou et al. Citation2020a; Frutos et al. Citation2021). In 2003, SARS-CoV pathogen emerged in bats and then transmitted to humans via intermediate host (civet cats) (Peiris et al. Citation2003). The direct interaction between animals and humans paves the way for adaptive mutation and interspecies transmission (Lu et al. Citation2015). Chronological reports of major CoVs outbreaks in humans and pigs have been illustrated in .
Figure 2. The chronological time-line of events presenting major coronaviruses outbreaks in swine (upper bar) and humans (lower bar).
Coronaviruses have posed devastating consequences on commercial animal production over the past few years, including infectious bronchitis virus (IBV) in poultry, bovine CoV as well as the emergence and re-emergence of multiple CoVs in swine such as TGEV, PEDV, and PDCoV. These animal CoVs infections facilitate understanding CoV disease as a whole; most of the animal CoVs have complex pathogenesis alike SARS-CoV-2 and thus can also throw light on the SARS-CoV-2 ongoing outbreaks amid COVID-19 pandemic and zoonotic concerns (Kenney et al. Citation2021). This review article presents an introductory overview on coronaviruses with a particular focus on swine coronaviruses (SCoVs) and their prevalence and epidemiology, virology, replication, evolution and emergence, clinical signs, pathology and pathogenesis, interspecies transmission, wildlife reservoirs, spill-over events and feasibility of the role of pigs in SARS-CoV-2 transmission during the ongoing pandemnic, and highlighting an overall scenario of emerging threats of SCoVs to swine population and human health.
2. Occurrence and epidemiology
Over decades, PRCV, TGEV and PHEV are prevalent in swine. While PDCoV, PEDV and SADS-CoV are emerging or re-emerging these CoVs pose a serious threat to pig farming. Chimeric PEDV and TGEV strains have also been documented in Italy, Germany, Slovakia, and Spain (Akimkin et al. Citation2016; Boniotti et al. Citation2016; Valko et al. Citation2019; de Nova et al. Citation2020). PEDV outbreaks were reported in Mexico, Czech Republic, Hungary, Belgium, China, Korea, Italy and Thailand during 1990s. It re-emerged as a major swine disease in China (Theuns et al. Citation2015; Niederwerder and Hesse Citation2018; Reveles-Felix et al. Citation2020). According to the most recent prevalence study in China, the average PEDV-positive rate in pigs from 39 farms across eight provinces, primarily in Hunan, was 38.0% (Tan et al. Citation2020). Later on, PEDV mutants were reported in Asian countries (Thailand, South Korea, Vietnam, and the Philippines) (Lee and Lee Citation2014; Cheun-Arom et al. Citation2016; Diep et al. Citation2018; Garcia et al. Citation2018), North America (Canada, Mexico) (Ojkic et al. Citation2015; Trujillo-Ortega et al. Citation2016) and in European countries (Austria, Ukraine, Belgium, Germany) (Steinrigl et al. Citation2015; Pensaert and Martelli Citation2016). Transmissible gastroenteritis virus and PEDV were co-circulating in Eurasia and the United States (Akimkin et al. Citation2016; Belsham et al. Citation2016; Mandelik et al. Citation2018). A pathogenic recombinant PEDV/TGEV mutant (swine enteric coronavirus, SeCoV) was found in Europe (Belsham et al. Citation2016; Boniotti et al. Citation2016). Even though, TGEV epidemiological studies are lacking in many countries (Valko et al. Citation2019). PRCV which is a TGEV mutation with a naturally occurring S gene deletion (170 to 190 kDa) has been reported in many European countries, including the Netherlands, Denmark, the United Kingdom, France and Spain (Magtoto et al. Citation2019; Turlewicz-Podbielska and Pomorska-Mól Citation2021). Porcine hemagglutinating encephalomyelitis virus, the only swine Betacoronavirus, is prevalent worldwide (Dong et al. Citation2014; Lorbach et al. 2017; Li et al. Citation2016). However, as per study in the United States of America, unusual respiratory (influenza-like) symptoms and the increasing prevalence of PHEV in adult swine could indicate a possible shift in their tropism, which could lead to a significant change in its epidemiology (Lorbach et al. Citation2017). Clinical outbreaks of vomiting and wasting disease (VWD) or PHEV-related fatalities were first reported in Canada in 1957 (Roe and Alexander Citation1958; Mora-Diaz et al. Citation2019). Later, clinical cases were recorded in many countries, including Argentina, the United States of America, Belgium, China, Japan, and South Korea (Gao et al. Citation2011; Dong et al. Citation2014; Mora-Diaz et al. Citation2019). During 2017 in Southern China, a unique bat-HKU2-like pig coronavirus (GDSO4 strain), i.e. porcine enteric Alphacoronavirus (PEAV), causing severe diarrhea in suckling piglets was reported (Gong et al. Citation2017). It was later known as swine acute diarrhea syndrome CoV (SADS-CoV) (Zhou et al. Citation2018).
Infection with PDCoV has been documented in several countries including the United States of America (Li et al. Citation2014; Wang et al. Citation2014; Ma et al. Citation2015), Canada, South Korea (Lee and Lee Citation2014; Lee et al. Citation2016), China (Liu et al. Citation2017; Li et al. Citation2018a; Zhang et al. Citation2019), Thailand (Madapong et al. Citation2016; Lorsirigool et al. Citation2017; Saeng-Chuto et al. Citation2017), Lao People’s Democratic Republic (Lorsirigool et al. Citation2016), Vietnam (Le et al. Citation2018), Mexico (Perez-Rivera et al. Citation2019), and Korea (Lee et al. Citation2016) causing serious threat to pig industry. In diarrheic pigs, the prevalence of PDCoV in China and the United States of America has been reported to be as high as 30 and 7%, respectively; and coinfections with PEDV are common in about 60% of cases (Zhang et al. Citation2019; Turlewicz-Podbielska and Pomorska-Mól Citation2021). Between 2015 and 2017, a study showed a prevalence rate of 36.2% of PDCoV in fecal samples from nine Chinese provinces (Ding et al. Citation2020). In Henan Province, China, researchers conducted a passive surveillance study to determine the evolutionary relationship between PDCoV strains. Phylogenetic analysis of the complete genome, nucleocapsid, and spike gene sequences showed that Chinese clade Henan strains of PDCoV were more closely related to other reference strains of PDCoV (Zhang et al. Citation2019).
Transmissible gastroenteritis virus (TGEV) was first reported in Belgium in 1984 and was first detected in the United States of America in 1946 (Doyle and Hutchings Citation1946; Pensaert et al. Citation1986). Cases of acute diarrhea in piglets in the United States of America led to the identification of TGEV in 1946 (Doyle and Hutchings Citation1946), and was later reported in Asia, Africa, South America and Europe resulting in significant economic losses in pig breeding sector (Kim et al. Citation2000; Stevenson et al. Citation2013; Cheng et al. Citation2020). Even though vaccines are widely used, TGEV infections are the primary source of enteric infection and mortality in piglets in the United States of America and around the globe from the 1960s to the 1980s (Yaeger et al. Citation2002). The existence and widespread occurrence of PRCV (deletion variant of TGEV), limited the therapeutic effect of TGEV (Turlewicz-Podbielska and Pomorska-Mól Citation2021). The enteropathogenic TGEV causes extreme diarrhea that is limited in swine. Even though pigs of every age group are prone to diseases, piglets exhibit the most severe clinical symptoms, with an almost 100% mortality rate. Due to development of resistance, the mortality rate decreases with age (Moon Citation1978; Turlewicz-Podbielska and Pomorska-Mól Citation2021). Piglets that survive from TGEV infection can recover within six to eight days, but virus can persist in lungs or gut upto 104 days and aid in the spreading of virus to non-infected pigs (Lawhorn Citation1999; Xia et al. Citation2018).
Unlike TGEV, PRCV has a preference for the respiratory system. A survey conducted in Belgium in 1984 led to the discovery of this virus. In the absence of vaccination, there was an increase in the number of animals with TGEV antibodies (up to 68%). Three years later, the virus had infected 100% of Belgium's swine farms (Enjuanes and Van der Zeijst Citation1995). The PRCV has been observed in a number of European countries, including the Netherlands, Denmark, the United Kingdom, Spain, and France (Enjuanes and Van der Zeijst 1995). Serological tests detected PRCV for the first time in Indiana, USA in 1989 (Enjuanes and Van der Zeijst 1995). Other PRCV isolates have since been reported in the USA and Canada (Enjuanes and Van der Zeijst 1995; Turlewicz-Podbielska and Pomorska-Mól Citation2021).
Porcine epidemic diarrhea virus (PEDV) was defined in island located at United Kingdom in 1977 (Niederwerder and Hesse Citation2018). In 1978, a ‘Coronavirus-like’ agent distinct from other identified SCoVs was reported (Chasey and Cartwright Citation1978). PEDV origin is unclear. There is no conclusive proof that bats introduced it into the swine farm in the 1970s. PEDV can replicate in cell lines from pigs, monkeys, bats, ducks, and humans, indicating that the virus has breached the interspecies boundary amongst bats and pigs (Liu et al. Citation2015; Teeravechyan et al. Citation2016). Porcine epidemic diarrhea virus was discovered in feral hogs in the Korean peninsula and the USA (Bevins et al. Citation2018). The virus had spread to most swine-producing areas within a year of its emergence, killing nearly 7 million pigs, mostly neonates in their first few weeks of existence. Porcine epidemic diarrhea virus has caused significant economic losses; with estimates of $300,000 in annual losses for a single 700-sow farrow-to-finishing herd (Weng et al. Citation2016). Infected pigs are the major source of infection to others, but it can also be spread through contaminated water or food, automobiles, shoes, and other inanimate objects. Workers involving in farm activities can similarly serve as vehicles to transmit viruses to newborn pigs (Dee et al. Citation2014, Citation2016; Schumacher et al. Citation2016). There has been some proof of PEDV transmission via aerogenous route (Alonso et al. Citation2014; Jung et al. Citation2020) but no other studies have described it till now. Currently, three PEDV genotypes are identified on the basis of S gene: I) G1a PEDV, which includes mildly virulent strains from Europe and Asia; ii) G2 PEDV, also known as ‘Original USA PEDV’, which includes highly lethal strains which was disseminated from Asia, presently prevalent in the USA; and iii) G1b PEDV, which embody S-INDEL strains (strains with insertions and deletions in the S gene linked to mild clinical outbreaks) (Decaro and Lorusso Citation2020). Infectious virus excretion up to 14–16 days in four-week-old pigs affected with an evolving PEDV strain (non-S INDEL) (Crawford et al. Citation2015).
Porcine hemagglutinating encephalomyelitis virus (PHEV) was first found in the brain of a suckling piglet suffering from encephalomyelitis and cultivated in primary porcine kidney (PK) cells (Greig et al. Citation1962). Porcine hemagglutinating encephalomyelitis virus was derived from bovine CoV, which is thought to have descended from a bat virus via rodent adaptation (Decaro et al. Citation2020). Porcine hemagglutinating encephalomyelitis virus has been the only neurotropic virus known to affect pigs, and it was one of the first swine coronavirus to be isolated. Later, a virus antigenically related to PHEV was identified in Europe as Von Willebrand disease (VWD) virus. Porcine hemagglutinating encephalomyelitis virus and VWD virus were first SCoV identified as a cause of encephalomyelitis (Mora-Diaz et al. Citation2019). Porcine hemagglutinating encephalomyelitis virus was recently linked to an influenza-like respiratory disease from pigs in Michigan, USA according to a new study (Lorbach et al. Citation2017).
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is an Alphacoronavirus that is most analogously related to coronavirus (HKU2) of bat, but is also related to HCoV NL63, HCoV 229E and PEDV (Gong et al. Citation2017). It was first identified and confirmed as the causal agents of a catastrophic pig epidemic in southern China in 2017, which killed nearly 24,500 piglets and resulted in significant commercial losses (Pan et al. Citation2017). Infection with SADS-CoV was linked to acute diarrhea and vomiting with mortality up to 90% in piglets under the age of five days (Zhou et al. Citation2019). Swine acute diarrhea syndrome coronavirus strain sequence resembles the HKU2 coronavirus of Rhinolophus spp. bats, implying that SADS-CoV may have originated from bats (Zhou et al. Citation2018).
Porcine Deltacoronavirus (PDCOV) is an emerging porcine enteric coronavirus which responsible for diarrhea in young pigs (Zhang et al. Citation2019) and reported in Hong Kong in 2012 (Woo et al. 2012). Porcine Deltacoronavirus has been shown to originate from sparrow CoV and known to be pathogenic to cattle and poultry apart from swine (Khamassi Khbou et al. Citation2021). Molecular clock analysis has shown that probable origin dated back to 1990s in South-east Asia with a common ancestry to sparrow CoV of around 1810 (Ye et al. Citation2020). But PDCoVs were previously identified in pigs and wild birds during preliminary investigations in China and Hong Kong from 2007 to 2011 (Boley et al. Citation2020). Yet, at the Chinese live-animal markets in 2005–2006 existence of PDCoV in small mammals like Asian leopard cats and Chinese ferret badgers were previously reported (Dong et al. Citation2007). Porcine Deltacoronavirus was found in diarrheic pigs on mainland China (Dong et al. Citation2015; Song et al. Citation2015). Interspecies transmission of PDCoVs is suspected amongst porcine, avian, and Asian carnivorous animals, because their S and helicase genes are closely linked to PDCoV (Li et al. Citation2018b). According to recent experimentation, PDCoV can cause infection and destroy cells from different animal and avian species (chicks and poults). Since avian susceptibility to PDCoV has been proven in vivo, but in vitro data indicating human susceptibility should be investigated further (Boley et al. Citation2020). The other swine enteric viruses including PEDV, TGEV and PDCoV infections cause vomition, diarrhea, dehydration and death in newborn piglets (Zhang Citation2016). In Ohio, USA the first epidemic of PDCoV-related diarrhea in pigs was recorded in 2014. Reverse trancription polymerase chain reaction (RT-PCR) testing revealed that 92.9% of diarrheic pigs intestinal or fecal samples from five Ohio farms were tested positive for PDCoV (Wang et al. Citation2014). In 2012, the first PDCoV strain was detected in Hong Kong during a CoV molecular epidemiology study that used RT-PCR to analyze pig samples. The complete genome sequencing revealed that the 2012 PDCoV strain sequences were very similar to the two Chinese prototype PDCoV strains (HKU15-155, HKU15-44) (Turlewicz-Podbielska and Pomorska-Mól Citation2021). Over the same time, two genetically related PDCoV strains (USA/IA/2014/8734 and SDCV/USA/Illinois121/2014) were identified in the USA (Li et al. Citation2014; Marthaler et al. Citation2014). Although this PDCoV has now been confirmed in 19 states of the USA (www.aphis.usda.gov/animal-health/secd); it is far less common than PEDV.
3. Genome and structure of swine coronaviruses (SCoVs)
Swine coronaviruses are single-stranded, polyadenylated, large genomic RNA (25–30 kDa) with positive sense polarity (. Similar genome profiles, replication strategies and protein expression are found in other animals’ and human CoVs (Chen et al. Citation2020). Swine coronaviruses are made up of four structural proteins: a large surface glycoprotein spike protein (S), an integral membrane glycoprotein (M), an envelope small membrane protein (E) and a nucleocapsid protein (N) (Song et al. Citation2019). Spike protein is a 180-kDa peplomer glycoprotein found on the virion envelope and the plasma membrane of infected cells. It contains epitopes for viral neutralization, diagnosis, and T-cell response to trigger immune response; and it is also required for virus cellular entry, as well as cell-to-cell fusion for some coronaviruses (Wang et al. Citation2020a). The spike gene contains factors of virus tropism and responsible for pathogenesis (Chen et al. Citation2020). Membrane (M) glycoprotein is a transmembrane protein with its carboxy-terminus embedded inside the virion core and it is thought to be essential in preserving the virion core structure (Kuo et al. Citation2016). The E protein is a membrane-associated polypeptide with a mass of 9.6 kDa that is essential for virus assembly (Castano-Rodriguez et al. Citation2018; Asrani et al. Citation2020). In addition, PHEV which is a beta-coronavirus contains hemagglutinin-esterase (HE) protein that forms second layer of shorter surface spikes (Turlewicz-Podbielska and Pomorska-Mól Citation2021). The nucleocapsid protein (N) with a mass of 60 kDa forms a helical structure of capsid inside the viral envelope by forming a complex with the genome RNA. The peplomers found in the envelope are shaped by spike protein trimers, giving the virions corona (crown-like) appearance (Wang et al. Citation2020a). The 3′-side of the genome encodes structural proteins such as E, N, S, M, HE and I 'internal protein' (encoded within the N gene) and HE (in certain beta-coronaviruses) as shown in . The replicase locus is located at the 5′-end of the genome it is made up of two large open reading frames (ORFs), i.e. ORF1a and ORF1b, which together occupy about 20 kb of the genome. The replicase is translated into two polyproteins (pp) 1a and 1ab. These replicase polyproteins are further cleaved by the virus proteases into 16 proteins, most of which contains proteases, multiple RNA modification enzymes, a helicase, and a polymerase (V'kovski et al. Citation2021).
Figure 3. Schematic diagram of swine coronavirus (SCoV) virion (S, Spike S structural gene; E, envelope; M, membrane; N, nucleoprotein; HE, hemagglutinin-esterase; Ns3a, Ns6, Ns7, Ns7a, Ns7b are accessory genes) (Mora-Diaz et al. Citation2021).
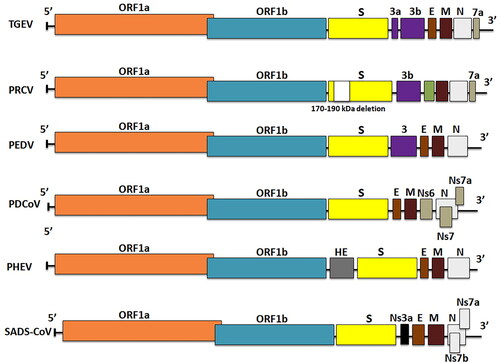
The TGEV genome is approximately 28.6 kb in size and consists of a 5′-end untranslated region (UTR), ORF1a/1b, S, M, E and N genes, and a 3′-UTR encoding three accessory genes such as 3a, 3 b, and 7 that are interspersed among the structural genes in the 3′-region (Chen et al. Citation2019). Cheng et al. (Citation2020) conducted a study that revealed two distinct genotypes of TGEV, genotypes I and II, with genotype I further classified into subtypes Ia and Ib using maximum likelihood and Bayesian inference phylogenetic analyses. In comparison to TGEV, the PRCV genome has a significant deletion (621 to 681 nucleotides) near the N-terminus, resulting in a smaller S protein, and some variable region deletion in the downstream of S gene, impedes ORF3 expression (Saif and Jung Citation2020). Porcine epidemic diarrhea virus has a genome of about 28 kb that comprises four structural proteins (S, E, M and N proteins), ORF3 as an accessory protein and sixteen nonstructural proteins (nsp1-nsp16) (Vlasova et al. Citation2020). The SADS-CoV genome is approximately 27 kb in length and includes 9 ORFs: ORF1a, ORF1b, S, E, M, N genes and three accessory genes, NS3a, NS7a and NS7b (Zhou et al. Citation2018). Porcine delta coronavirus has a 25.4 kb genome that encodes 4 structural proteins [S, M, E and N], along with three accessory proteins (NS6, NS7 and NS7a) (Li et al. Citation2014; Wang et al. Citation2019). NS6 is anticipated to exist between M and N in the genome and it has recently been discovered as a virion-associated inhibitor of IFN-expression and has been proven to suppress the host innate immune response (), but NS7 is a 200-amino-acid peptide encoded by an alternate ORF within the N gene (Fang et al. Citation2017). Modulating viral pathogenicity, inducing apoptosis, and antagonising the IFN system are a few roles of these accessory proteins (Koonpaew et al. Citation2019). A previous phylogenetic analysis based on spike genes suggested that PDCoV was classified into three groups: China, the USA, and Southeast Asia (SEA). Southeast Asia PDCoV, is genetically distinct from China and the USA, but they share a common ancestor. The emergence and evolution of SEA PDCoV were explored using phylodynamic analysis methods based on complete genome. In the study, 18 complete genome sequences of SEA PDCoV identified between 2013 and 2016, as well as PDCoV from other regions, were analysed. The findings revealed that PDCoV was classified into two genogroups, G1 and G2. G1 is further subdivided into G1a (China), G1b, and G1c (USA). The G2 (SEA) group has further evolved into three clades: SEA-1 (Thailand), SEA-2 (Vietnam), and SEA-2r (Vietnam recombinant) (Stott et al. Citation2021).
3.1. Coronavirus replication
Coronavirus replication begins with interaction between the S protein and receptor for the viral entry into the host cell. Coronaviruses' host species range and tissue tropism are largely controlled by this interaction. The relatively variable S1 subunit mediates receptor binding, while the more conserved S2 subunit undergoes large conformational changes, resulting in virion and cell membrane fusion. The outer membrane, or in some cases, acidified endosomes are the sites of the membrane fusion (White and Whittaker Citation2016). Different CoVs in nature use diverse range of receptor to get entry inside the cell. Different SCoVs tropism and receptors are summarized in . In conjunction to receptor binding and fusion, coronavirus S proteins should be proteolytically cleaved by host cell-derived proteases (V'kovski et al. Citation2021). After genomic RNA uncoating and cytoplasmic entry, the RNA instantaneously translates the two large open reading frames, ORF1a and ORF1b into two polyproteins, pp1a and pp1ab. The specific non-structural proteins (nsps) that support the virus replication and transcription complex are generated from the polyproteins (pp1a and pp1ab) through cis- and trans-cleavage of the polyproteins by the virus proteases (V'kovski et al. Citation2021). After that, several of the nsps constructs to form replicase–transcriptase complex (RTC), that further develops environment to synthesize RNA, including transcription and replication of subgenomic RNAs (Wang et al. Citation2020a).
Table 1. Different swinecoronaviruses, tropism, and their receptors.
The translation and assembly of viral replicase complexes are followed by synthesis of viral RNA including genomic and subgenomic RNAs. The order Nidovirales is having unique properties of nested set of transcription for production of subgenomic RNAs. Subgenomic RNAs work as mRNAs for the synthesis of structural and auxiliary/accessory genes. All positive sense subgenomic RNAs are from 3′ co-terminal which is independently transcribed, forming a set of nested RNAs. Both genomic and subgenomic RNAs are generated through negative-strand intermediates (Wang et al. Citation2020a). Each mRNA is translated to produce the protein product of its most 5′-gene, but it can also be translated to produce a second, downstream protein. It has a mysterious discontinuous transcription mechanism that allows them to replicate. Transcription regulating sequences (TRSs) at the 5′-terminal of each transcriptional unit are required to regulate subgenomic mRNA transcription (Du Toit Citation2020). The translation and incorporation of virus structural proteins S, E and M into the cell organelles such as endoplasmic reticulum occurs immediately after viral replication. These proteins find their path into the vesicular tubular cluster (VTC) through the secretory pathway (Wang et al. Citation2020a). However, viral genomes encapsidated by N protein bud into membranes of the VTC encompassing viral structural proteins, creating complete virions (V'kovski et al. Citation2021). The M protein guides most protein–protein interactions obligatory for assembly of CoVs. Subsequently, new virions bud from intracellular membranes and then excluded out from the cells through secretory vesicles by the process of exocytosis (Chen et al. Citation2020). Swine CoVs are considered difficult to propagate. However, certain studies like the use of embryonated chicken eggs have been utilized successfully to propagate deltacoronaviruses which can be helpful for vaccine production and isolation of coronaviruses (Iseki et al. Citation2021).
4. Coronavirus evolution
Novel viruses require access to new hosts, which is often achieved through ecological disruption and the ability to efficiently infect the hosts, which is frequently driven by adaptive evolution. Coronaviruses can quickly adapt to new hosts by using genetic recombination and mutation. A new virus cannot be created solely through point mutations but when the same host is infected with two coronavirus strains at the same time, leads to the recombination of hundreds or thousands of base pairs in long genomic fragments, resulting in the emergence of new virus (Trovato et al. Citation2020). Three new human and swine CoVs have emerged and expanded globally in the 21st century, highlighting the critical need of detection techniques for zoonotic coronaviruses (Wang et al. Citation2019). Mutation and recombination are the two major forces that drive coronavirus evolution. Coronaviruses have an increased frequency of homologous RNA recombination, but the mechanism is unclear. Evolutionary changes like mutation and recombination along with genetic drift and pleiotropy have a significant impact driving viral adaptations to various hosts. Phylogenetic studies have demonstrated evolutionary changes in the S glycoprotein of bovine coronavirus (BCoV) and human coronaviruses (hCoVs), and these modifications do not alter the viruses to retain in population, but it also influences the pathogenicity and the virulence (Miller Citation2005; Ren et al. Citation2006; Bidokhti et al. Citation2013; Lee and Lee Citation2018). Mutation in viruses induces the increased efficiency to bind with the host receptors, and some mutation occurs to evade the host immune response and assure continued dissemination by the host for successful transmission to another variety of animal (Oberemok et al. Citation2020). Viral cross-species transmission and infection rely on fast evolution, which is achieved through high genetic changes, recombination, along with the proximity of potential host species (Wu and Kewal Ramani Citation2006; Streicker et al. Citation2010; Lu et al. Citation2015; Abdullahi et al. Citation2020). Pathogenicity and dissemination capacity are increased by mutations that enable faster and efficient host entrance, maximize viral reproduction, and reduce sensitivity to host immune responses, either inside the host species or even across species (Longdon et al. Citation2014; Ahmed et al. Citation2020; Brook et al. Citation2020). Earlier cross-species transmission investigations have confirmed the necessity of monitoring genetic changes of virus in the new host. For example, in SARS-CoV-2, variations to the receptor-binding domain (RBD) modify virus infectivity, virulence and natural immunity or vaccination effectiveness (Korber et al. Citation2020).
Coronavirus (CoV) evolution is further controlled by genetic variation, viral mutation potential, and genetic recombination, along with surface glycoprotein adaptability, which is typically enhanced via host-parasite contact despite decreasing pathogenicity (Alexander and Day Citation2010; Alnazawi et al. Citation2017). On the other hand, polymorphisms in particular genes not just affect the gene's potential morphological expression but can also impact additional genetic traits and subsequent transcriptional activities of other genetic sequences (Bang Citation1978; Van Sluijs et al. Citation2017; Lee and Lee Citation2018). Coronavirus genome recombination can be aided by signals for RNA polymerase recognition. Recombination has only been confirmed between CoVs of the same genus, despite the possibility of intergroup recombination. Intragroup recombination has been reported between canine and feline enteric coronaviruses, mouse hepatitis virus (MHV) and infectious bronchitis virus (IBV) strains (Su et al. Citation2016). During coinfection, a polymerase-jumping system causes recombination among coronaviruses (Su et al. Citation2016). In cell culture, recombination between human (HCoV-OC43) and animal (bovine) CoVs has recently been reported. It has been proposed that members of group II CoVs like hemagglutinating encephalitis virus (HEV) have a high capacity for recombination (Wu et al. Citation2003). RNA viruses have been observed to mutate at frequencies ranging from 103 to 105 base substitutions per nucleotide (Duffy Citation2018). These values are higher than those seen during the replication of DNA virus and cellular DNA (Vlok et al. Citation2019). RNA viruses form into diverse populations consisting of ensembles of highly related but non-identical genomes referred as viral quasispecies as a result of their high mutational rate (Domingo and Perales Citation2019). The minimal copying fidelity shown by viral replicases is the molecular basis of all this uncertainty (Kautz and Forrester Citation2018).
Swine acute diarrhea syndrome (SADS) has recently been detected in piglets and it is caused by the SADS coronavirus (SADS-CoV), a new strain of coronavirus HKU2 from Rhinolophus bat (Zhou et al. Citation2018), but till now there has been no evidence of human infection. In the case of PDCoV, molecular studies indicate that interspecies transmission occurred recently as a result of contact between bird and mammal (Ma et al. Citation2016) and also virus progenitors were found among quails and sparrows (Li et al. Citation2018c). A recent report of finding porcine deltacoronavirus strains in three Haitian children’s serum samples with undiagnosed febrile illness also raises a question of interspecies transmission of other CoVs as till now humans get infected with only alphacoronaviruses and betacoronaviruses (Lednicky et al. 2021a). In fact, the broad cross reactivity of deltacoronaviruses with cellular receptor–aminopeptidase N (APN)–from different species also supports this kind of evolution (Vlasova et al. Citation2021).
The virus cannot evolve in isolation but is instead complemented by changes in the host as well. Host evolution and ecology, linked to genetic changes and recombination processes, offer the essential host variables that must occur concurrently with virus evolution (Fumagalli et al. Citation2010; Cui et al. Citation2013; Nasir et al. Citation2015; Lee and Lee Citation2018). Several variables must come along for successful zoonoses and the capacity to build in a new host, along with the development of a founder virus with a modification that enables higher levels of dissemination in the new host (Jelinek et al. Citation2021).
4.1. Recombination
Coronaviruses are experts at RNA recombination, a genetic process in which two separate viral genomes of RNA hybridize and swap RNA regions at specific locations. RNA recombination occurs frequently in the genomes of CoVs during replication and thereby integrity of RNA is maintained. Coronaviruses can also acquire new genes, which allows them to accommodate to a new host. Variations in other regions of the CoV genome can either make it more adaptable to the new host, or the genome can incorporate host/cellular RNA that codes for an accessory component that enables it to replicate in a different species. As a result, it is clearer than ever that new CoVs pose a significant risk (Alluwaimi et al. Citation2020). RNA recombination events that result in mutations of S protein are classic explanations of how viral genomic modifications enable a coronavirus to evolve and bind to a target ligand, allowing it to infect a new species (Ji et al. Citation2020).
Due to the high degree of mutation that defines their coexistence in a diverse range of variants, CoVs appear as quasispecies (Mandary et al. Citation2019). CoV RNA mutation rates have been reported to be moderate to high. The estimated annual substitution rate for CoVs was 104 substitutions per site. For example, the IBV hypervariable area of the S gene's nucleotide mutation level was determined to be 0.3–0.6 × 102 per site annually. In 229E CoV, the rate of substitution was determined to be ∼ 3 × 104 per site per year, while in SARS-CoV, the rate was determined to be 0.8–2.38 × 103 nucleotides per site annually (Su et al. Citation2016). As shown during the SARS outbreaks in 2002–2003, the phylogeny and biology of the coronavirus (CoV) was likely characterized by frequent host-shifting events, whether animal-to-human (zoonosis) or human-to-animal (reverse zoonosis) (Lau et al. Citation2018). SARS-CoV has shown evidence of recombination with PEDV at some point of evolutionary history (Su et al. Citation2016).
4.2. Interspecies transmission
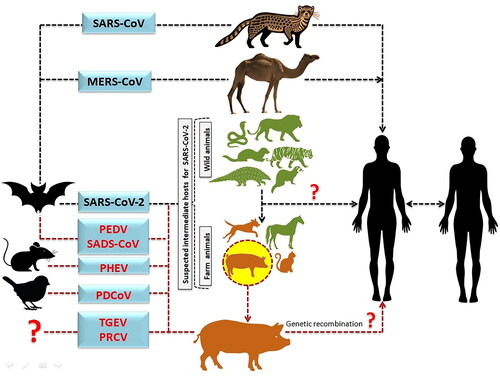
The definition ‘Species barrier’ refers to natural mechanisms that prevent pathogens from spreading from one species to another. Spill-over is the transition of an organism from a host to another recipient species, as shown in . Pathogens can propagate in a variety of ways, including from severely diseased domestic animal hosts to non-domestic hosts (Borremans et al. Citation2019). A complex pathogen transmission is being created by these pathogens spill-over and emergence events. Apart from the dietary and cultural factors, the ecological, epidemiological and human inherent factors may also link with spill-over of food-borne zoonotic diseases (Dhama et al. Citation2013a; Dhama et al., 2013b; Dhama et al., 2013c; Iturriza-Gomara and O'Brien Citation2016; Plowright et al. Citation2017). The interaction of viral-specific S protein, with the host ACE2 receptor supports spillover of CoV (Salata et al. Citation2019; ; Tiwari et al. Citation2020). Mutations in receptor binding domain (RBD) of S protein cause changes in receptor specificity, interaction and binding affinity, resulting in changing of transmissibility, pathogenicity, and cross-species jumping, as well as a predisposition to latest and more catastrophic diseases (; Wang et al. Citation2020c). Before 2002, research showed that mammalian CoVs may infect birds and cross-species barriers. Even though poultry are vulnerable, chicks and pigs were not susceptible to bovine coronavirus infection (Ismail et al. Citation2001; Saif and Jung Citation2020).
Figure 4. Diagrammatic depiction of the source of SARS-CoV-2 from bats and presumed intermediate hosts, such as farm and wild animals. (The transmission of MERS-CoV and SARS-CoV to humans via an intermediate host camel and palm civet cat, respectively. It also illustrates the likelihood of SARS-CoV-2 originating from pigs after genetic recombination, which would enable the SARS-CoV-2 to cross the species boundary and transmit disease to humans.)
Experiments have shown that numerous subgroups of CoVs, such as canine enteric CoV (CECoV) and feline CoV (FCoV), are strongly linked to the TGEV/PRCV cluster, with interspecies transmission. All of these viruses are closely related biologically, antigenically, and genetically, and they could be host range variant of a CoV (Saade et al. Citation2020). They cross-react with monoclonal antibodies (mAbs) targeting to the S, N, or M protein in virus neutralization tests. They all share identical antigenic sub-site A on the S protein. Interestingly, antibody-dependent enhancement (ADE) of FCoV (feline infectious peritonitis virus [FIPV] strains) infection on macrophages cell culture is efficiently facilitated by neutralizing immunoglobulin G (IgG) mAbs against S protein of TGEV, illustrating the functional cross-reactivity of the S antibodies. The N and M proteins of type II FCoV, CECoV, and TGEV share >90% identical amino acids and the S protein shares >80% lengthwise identical amino acids (Saif and Jung Citation2020). Aminopeptidase N is a cellular receptor shared by TGEV, PRCV, and FCoV. The cross infection can occur between pigs, dogs and cats, causing symptoms such as profound blood-spattered diarrhea of which its occurrence remains mysterious (Saif and Sestak Citation2006). Amino acid composition of APN receptor in human, feline, and swine CoVs show 78% identity. The APN receptor used by alpha CoVs, on the other hand, is species-specific, and the feline APN is a functional receptor for other apha CoVs, including TGEV. Because of the characteristics of feline APN, cats can get infected with TGEV with or without symptoms (Parkhe and Verma Citation2021). A chimeric virus with a PEDV spike protein and TGEV backbone was identified as a variant genotype of TGEV strain with unique deletions and distinct amino acid changes similar to PRCV, implying recombination between the variant TGEV, PEDV, and PRCV (Belsham et al. Citation2016; Boniotti et al. Citation2016). Lednicky et al. (2021b) discovered porcine deltacoronavirus strains in plasma samples from three Haitian children suffering from acute undifferentiated febrile illness. Human infections were caused by at least two distinct zoonoses with the same mutational signature in the genes encoding Nsp15 and the spike glycoprotein, according to genomic and evolutionary analyses. Bat CoVs have a broad range as well as a great potential for spill-over in multiple animal species. As previously reported in the civet cat and dromedary camel, which culminated in the well-known catastrophes SARS and MERS, respectively, whereas SADS is latest spill-over in pigs which showing 95% genome similarity with bat CoV and causing high mortality (24,693 death) in neonatal piglets (Zhou et al. Citation2018).
5. Porcine CoVs: pathogenesis and clinical signs
5.1. Transmissible gastroenteritis virus (TGEV)
The most frequent clinical manifestation in piglets is vomiting, accompanied by milky yellow diarrhea, dehydration, and weight loss. Death may occur in 2–7 days after the onset of illness in animals less than the age of 7 days. In-appetence and diarrhea last for a shorter period in older pigs, but subclinical infections can often be seen. Extreme symptoms in lactating sows include pyrexia, vomiting, diarrhea, and failure of sow to produce milk. The time-period for the disease to develop can last up from 18 hours to 3 days (Mostl Citation1990). Xia et al. (Citation2018) reported endemic form generally lasts for short period of time, but in susceptible animals, it may remain in an enzootic form most of the time. The majority of sows in these herds become infected and their suckling piglets develop passive immunity. In such circumstances, diarrhea is more likely to occur after weaning (Zhang et al. Citation2017; Xia et al. Citation2018). Gross pathology is limited to the gastrointestinal tract that includes a bloated stomach with fluid-filled intestine which having undigested milk, gas and revealed flabby consistency on palpation. Damage to the villous enterocytes causes thinning of the intestinal wall which can be seen when segments of the intestine are immersed in an isotonic buffer and examined underneath a dissecting microscope. The villous atrophy is marked in jejunum and to some extent in ileum but not in proximal duodenum (Guo et al. Citation2020). Xia et al. (Citation2018) observed that newborn pigs had severe villous atrophy compared to 3-week-old pigs, implying that neonates are more prone to more severe TGEV infection. The functions of villi are quickly restored after recovery since the intestinal crypt epithelial cells continue to regenerate (Fenner Citation2017). The mammary gland tissues and alveolar macrophages have been identified as extra-intestinal sites for replication of TGEV (Vlasova et al. Citation2020). Transmissible gastroenteritis virus induced pyroptosis in porcine intestinal cell line and produce pro-IL-1β by caspase-1 activation (Wei et al. Citation2021). Porcine kidley-15 (PK-15) cell lines are preferred to isolate TGEV causing cytopathic effect (CPR)- like cell fusion, cell rounding, shrinkage, and cell detachment from the surface of the culture vessel (Yuan et al. Citation2021).
5.2. Porcine respiratory coronavirus (PRCV)
Patho-morphological study of PRCV revealed macroscopic and microscopic lesions which are typically confined to the lungs and manifest as lung consolidation, bronchointerstitial pneumonia with recurrent peribronchiolar and perivascular lymphohistiocytic cuffing (Cui et al. Citation2020). The similarity of nucleotide and amino acid sequences between TGEV and PRCV genomes is 96–98%, implying that PRCV originated from TGEV. Two aspects of the PRCV genome that may enlighten its altered tissue tropism which are a significant exclusion (621–681 nt) in the 5′ of the S gene, resulting in a smaller S protein, and in constant sequence alterations of ORF3 gene (Saif and Jung Citation2020).
Porcine respiratory coronavirus has a tropism for both the upper and lower airway passage similar to SARS-CoV and SARS-CoV-2. Similar to PRCV in pigs, most individuals infected by SARS-CoV-2 or SARS-CoV has an asymptomatic or subclinical infection and recuperates. Respiratory disease can be persistent in seriously affected patients, and it can be exacerbated by a storm of cytokines and multi-organ failure. In a non-human primate model, SARS-CoV-2 and SARS-CoV caused extreme diffuse alveolar damage and pulmonary edema due to excessive degeneration of type 1 pneumocytes of lung alveoli (Rockx et al. Citation2020). For PRCV diagnosis and differentiation from TGEV, real-time reverse transcription polymerase chain reaction (qRT-PCR) was used (Saif and Jung Citation2020). Using swine testicle and pig kidney (PK) cell lines, PRCV was isolated from nasal swab fluids or lung tissue homogenates and propagated in cell culture (Saif et al. Citation2019). Porcine respiratory coronavirus serology is complicated owing to cross-reactivity with TGEV.
5.3. Porcine epidemic diarrhea virus (PEDV)
The PEDV pathogenesis is more related to that of TGEV. The destruction of epithelial cells in the small and large intestine causes villous shortening. Viral replication takes place in the cytoplasm of epithelial cells of villi over the small intestine within 12–18 hours of post infection (PI) and a maximum effect was reached at 24–36 hours. Enterocyte degeneration occurs as a result of the infection, lowering the ratio of intestinal villous to crypt from 7:1 to 3:1 (Jung et al. Citation2020). Clinical manifestation of diarrhea, vomiting along with dehydration occurs after a 22–48-hour incubation period and is difficult to distinguish from TGEV symptoms. Unlike TGEV, the most affected groups are feeder and weaned pigs (Wu et al. Citation2020). In suckling piglets, PEDV can cause serious clinical disease and mortality may reach approximately to 80–100%. TGEV propagate more rapidly in diseased herds, resulting in a gratifying serious syndrome, but Weng et al. (Citation2016) indicated that TGEV cases in the previous year have shown an altered clinical pattern, resembling PEDV. Porcine epidemic diarrhea virus and TGEV have very close pathogenetic mechanisms that result in the same immunological condition. Recent studies also showed increased PEDV pathogenicity on co-infection with porcine delta-coronavirus (Zhang et al. 2021c). A study also showed the RBCs as a possible vehicle for transmission of PEDV which could also be possible in other coronaviruses (Li et al. Citation2021). Immunity to PEDV is dependent mostly on secretory antibody in mucosa of small intestine, which is only present for a brief period of time following disease. Antibody transfer through milk protects suckling piglets, but circulatory antibodies do not. To diagnose PEDV, blocking-ELISA is mainly used to detect viral antigen or by an immunofluorescence assay (Magtoto et al. Citation2019). El-Tholoth et al. (Citation2021) designed a new, low-cost, rapid, semi-quantitative, field deployable, 3 D-printed microfluidic device for auto-distribution of samples and self-sealing, as well as real-time and reverse transcription-loop-mediated isothermal amplification (RT-LAMP), allowing the co-identification of PEDV, TGEV, and PDCoV in around 30 minutes.
5.4. Porcine hemagglutinating encephalomyelitis virus (PHEV), vomiting and wasting disease (VWD)
The virus would be particularly transmitted through nasal secretions. In pigs, the respiratory tract is the primary site for PHEV replication. The virus replication occurs in the interior of the upper respiratory tract epithelium without inflicting clinical symptoms in majority of cases. Afterwards the virus moves towards the brain and spinal cord through peripheral nerves, resulting in encephalomyelitis or VWD. Several pathways were used by the virus to disseminate from the initial replication sites to the CNS via peripheral nervous system. One pathway connected the nasal mucosa and tonsils to the trigeminal ganglion and trigeminal sensory nucleus in the brain stem. The vagal sensory ganglion provided a second route from the vagal nerves to the vagal sensory nucleus in the brain stem. Following replication in local sensory ganglia, a third pathway reaches the spinal cord from the intestinal plexi (Mora-Diaz et al. Citation2019). Almost all piglets under the age of three weeks show severe clinical signs. They vomit frequently, pale, listless and huddle together after a 4–7 day-incubation period. Mora-Diaz et al. (Citation2021) proposed that vomiting is due to virus replication within infected neurons or through vagal sensory ganglion in the vomiting center. Widespread muscle twitching and hyperesthesia are typical symptoms of encephalomyelitis but opisthotonus, blindness and nystagmus may also occur. Weakness is followed by syncope and collapse in the large number of piglets. An interruption in stomach emptying was thought to be the cause of the wasting (Mora-Diaz et al. Citation2019).
Even though PHEV is found in pig populations all over the world, clinical disease is rare. Due to the fact that the majority of pigs are seropositive, their young ones are shielded by maternal antibodies until they acquire age-dependent protection. Asymptomatic infections develop in pigs between ages of 8 and 16 weeks throughout the passive immunity phase, inducing active immunity (Mora-Diaz et al. Citation2021). Cachexia, stomach dilatation and abdominal distension have been reported as the only gross lesions in some chronically infected pigs with natural PHEV infections (Mora-Diaz et al. Citation2020). Histopathological lesions such as epithelial degeneration and infiltration of inflammatory cells are visible in the tonsils and respiratory tracts of severely diseased pigs. Non-suppurative encephalomyelitis was reported in 70–100% of the pigs with nervous symptoms and 20–60% of the pigs with VWD syndrome. Gliosis, perivascular cuffing and neuronal degeneration are the characteristic lesions (Mora-Diaz et al. Citation2019). Porcine hemagglutinating encephalitis virus is similarly interrelated to bovine, canine, murine, equine and human CoVs, accompanied by rat sialolodacryoadenitis coronaviruses (Mora-Diaz et al. Citation2019). Porcine hemagglutinating encephalitis virus has an envelope-associated glycoprotein layer (hemagglutinin-esterase) that allows it to hemagglutinate and hemadsorb chicken, mouse, hamster, rat, and turkey erythrocytes, that distinguishes it from other swine coronaviruses (Sasseville et al. Citation2002; Mora-Diaz et al. Citation2021).
5.5. Swine acute diarrhea syndrome coronavirus (SADS-CoV)
In Guangdong province, China, the swine enteric Alphacoronavirus (SeACoV) was identified in 2017. It is also known as SADS-CoV and porcine enteric Alphacoronavirus (PEAV). Initially, epidemics of profound diarrhea in nursing piglets were reported at four pig farms in a hilly region of north Guangdong (Gong et al. Citation2017; Zhou et al. Citation2018). Swine acute diarrhea syndrome coronavirus was first recovered in the fecal material of young pigs with severe diarrhea as well as an increase in fatality rate (Pan et al. Citation2017). Virus resistance and species tropism were also studied in cell lines derived from tissues of humans and several animal species, including bats, mice, rats, gerbils, hamsters, chickens, pigs, and nonhuman primates.
Swine acute diarrhea syndrome coronavirus, like other pigs enteropathogenic viruses, is spread via the fecal-oral route. This was shown in pigs using cell-cultured SADS-CoV strains during challenge trials (Pan et al. Citation2017; Zhou et al. Citation2018; Xu et al. Citation2019). Swine acute diarrhea syndrome coronavirus is most likely spread through vomitus or diarrheic stool from diseased piglets. Other PCoVs may be capable of infecting pigs in a number of ways. For instance, PEDV can cause intestinal mucosal infection if it enters in the body through the nose (Li et al. Citation2018b). Future studies should also focus on the infectious nature of aerosolized SeACoV.
SADS-CoV could produce a similar public health risk if it is transmitted from pigs or other intermediate hosts to human (Yang et al. Citation2019). Based on recent molecular studies, SADS-CoV has been discovered to have the potential for further cross-species transmission, including the ability to infect primary human airway, fibroblasts, human nasal epithelial, microvascular endothelial cells (MVE), and intestinal cells (Goldstein et al. Citation2021). In vitro experiments showed that SADS-CoV can grow effectively in monkey and nine human cell lines, suggesting that it has zoonotic potential. Since a conserved receptor for SADS-CoV cell entry has been proposed across various animal species, via interspecies transmission from porcine, SADS-CoV may pose a potential threat to human health in the future. Similar to other porcine enteric CoVs, SADS-CoV infects intestinal epithelial cells as its primary target cells (Koonpaew et al. Citation2019; Wang et al. Citation2019). Studies reported that in the intestine of SADS-CoV infected piglets proclaimed significant atrophy of villi causing devastating pathological lesions (Zhou et al. Citation2018; Koonpaew et al. Citation2019; Wang et al. Citation2019; Xu et al. Citation2019). The pathophysiology of SADS-CoV and the natural resistance to it are either poorly understood or conflicting. More investigations into the pathogenesis of this disease are required.
5.6. Porcine deltacoronavirus (PDCoV)
Although the etymology of PDCoV in USA swine is unidentified, serologic data show that it was propagating in swine prior to its discovery in 2014 (Sinha et al. Citation2015). Porcine Deltacoronavirus has also been identified in Canada (March 2014), Korea (April 2014), Thailand (2015), mainland China (2015), Vietnam and the Lao PDR. According to a study in Korea, only two samples from one farm were PDCoV RNA positive out of 691 diarrheic pig fecal samples collected from 2014 to 2015 (Lee et al. Citation2016). The SL2 and SL5 Korean PDCoV strains were genetically related to the USA PDCoV strains, whereas they were not related to the older Korean strain, KNU14-04. There was a high occurrence of PDCoV (>30%) and periodic co-infections with PEDV (51%) reported. The global PDCoV strains and Chinese PDCoV strains shared a high nucleotide identity which is around 98.9%. The Thai PDCoV strains had the highest nucleotide identity (98.4%) with the Chinese strain CHN-AH-2004 (Janetanakit et al. Citation2016), but they formed a different cluster from the Chinese and USA strains (Zhang Citation2016). Strains of the Thai PDCoV lineage were detected in Lao PDR, while strains of the USA PDCoV lineage were identified in Vietnam (Saeng-Chuto et al. Citation2017). Recent studies have established four distinct phylogenetic lineages of PDCoV (USA, China, Thailand, and Early China lineages); each with its geographic distribution pattern (He et al. Citation2020). In piglets, PDCoV causes diarrhea, vomition, dehydration with typical histopathological lesions of atrophic enteritis (Zhang et al. Citation2019). The fecal-oral route is the most common way for PDCoV transmission. The infection is spread mostly by feces, blood, and other infected fomites. The duration of experimentally produced PDCoV diarrhea was 5–10 days, and fecal viral shedding persists upto 19 days (Hu et al. Citation2016). After recovering from an infection, pigs mostly tend to shed PDCoV in their feces. Hence, subclinically infected or convalescent carriers may be another potential reservoir for PDCoV.
Porcine Deltacoronavirus is having hemagglutinating (HA) property which has been documented by Zhang et al. Citation2019. The virus had no HA reactivity for the pig, guinea pig, mouse, chicken or human erythrocytes, but it can cause agglutination of rabbit red blood cells when pre-treated with neuraminidase or trypsin. Furthermore, the results of the HA assay exhibited a positive relationship with the infectious viral titer. The findings show that assessing PDCoV HA activity may be a promising diagnostic tool for inspecting and monitoring PDCoV disease in pig population (Zhang et al. Citation2019).
6. Could pig be a vector for SARS-CoV-2?
Pigs are vulnerable to a variety of CoVs, including TGEV, PEDV, PRCV and the newly discovered SADS-CoV (Pan et al. Citation2017; Zhou et al. Citation2018). These viruses seem to have emerged in bats, similar to SARS-CoV and MERS-CoV. Pigs are also vulnerable to a Betacoronavirus, PHEV (Mora-Diaz et al. Citation2019), Deltacoronavirus CoV, and PDCoV (Zhang et al. Citation2019). As stated earlier, pigs and humans have distinct ranges of CoVs that do not overlap significantly. It is crucial to highlight that CoVs primarily affects the respiratory system in humans and enteric system in pigs. Coronaviruses have a very high mutation rate since they are RNA viruses, which help them to produce new strains/variants and adapt to a wide variety range of hosts. As a result, all identified CoVs of human have animal origins, according to genome sequences (Cui et al. Citation2019). In pigs, only one CoV is related to respiratory infections (PRCV), which causes mild disease and lesions in majority of cases (Cui et al. Citation2020) and is thus of little interest to the farmers. Only a few studies have looked at whether SARS-CoV or MERS-CoV can infect pigs, and whether pig viruses can infect humans. RNA of SARS-CoV was found in one pig during a survey of 6 domestic animal species, which also includes pigs, and 242 individual pigs in China (Opriessnig and Huang Citation2020). Also, two of the 242 pigs tested antibody positive for SARS-CoV. According to the researchers, interspecies transmission of SARS-CoV is possible and could pose a threat to human population (Opriessnig and Huang Citation2020).
Moreover, SARS-CoV replication was also supported by successive cultures of virus in turbinate cells of pigs (PT-K75) (Weingartl et al. Citation2004). MERS-CoV has also been shown to be amplified in pigs. Currently, there is no proof that pigs can be diagnosed with SARS-CoV-2 or they can spread the virus. Since the SARS-CoV-2 infection is still underway, all testing at this point focuses on detecting affected individuals to avoid more human-to-human transmissions. Early pandemic warnings were focused on pigs because they have been known to incubate other viruses, such as influenza, and they live in large numbers in close vicinity to humans in China, the place where the pandemic began, where 300 million pigs were farmed (Mallapaty Citation2021). SARS-CoV-2 replication was found to be successful in cats and ferrets but ineffective in pigs, rats, chickens, and ducks in an experimental infection study (Shi et al. Citation2020).
Pigs are generally not found susceptible to SARS-CoV-2 and may tend to have a low susceptibility to viral infection, requiring a very high infectious dosage to elicit a low degree of infection (Schlottau et al. Citation2020; Younes et al. Citation2021). In one experimental study, SARS-CoV-2-inoculated domestic pigs showed no evidence of clinical signs, virus replication or virus-specific antibody responses, thus were not found susceptible to the viral infection on oral/intranasal/intratracheal challenge, revealing that pigs are unlikely to be potential carriers of SARS-CoV-2 and neither to be considered a suitable animal model for SARS-CoV-2 (Meekins et al. Citation2020). Certain pig breeds like Ossabaw pig can be developed as obese and could present nearly all features of the metabolic syndrome, thus resembling to more than 80% of the critically ill COVID-19 patients, hence infection with porcine respiratory coronavirus of metabolic syndrome pigs (obese Ossabaw pig) has been suggested to considered as a highly relevant animal model of severe COVID-19 (Heegaard et al. Citation2020).
Pigs are most likely to contract SARS-CoV-2 because they have the same ACE2 receptor binding sites as humans (Wan et al. Citation2020). Several lines of evidence suggest that pigs could be prone to SARS-CoV-2 infection, for both natural and laboratory environments, and exhibit seroconversion. SARS-CoV-2 can use the ACE2 receptor from animal species, including porcine ACE2, to enter the cell in vitro, implying that pigs may be vulnerable to the infection of SARS-CoV-2 (Zhou et al. Citation2020b). Evaluating ACE2 receptor-mediated SARS-CoV-2 entry across different animal species, a key determinant for determining the range of hosts that could be infected by this pandemic virus, it was observed that ACE2 of Sus scrofa (wild pig) facilitated virus entry into non-susceptible cells (Zhang et al. Citation2021c). Receptor binding domain of spike protein of SARS-CoV-2 links with the pig ACE2 entry receptor with comparable efficiency to ACE2 entry receptor of human, as per structure-based observations (Wan et al. Citation2020). Pigs will co-express entry receptor ACE2 and TMPRSS2 protease as a viral activating factor in different cell lines, according to single-cell screening, and SARS-CoV-2 replicates in different cell lines of pigs (Chu et al. 2020; Schlottau et al. Citation2020; Zhou et al. Citation2020b). SARS-CoV-2 was able to replicate in two different porcine cell lines, viz., swine testicle (ST) and porcine kidney (PK-15) cell lines that were found permissive for the virus, inducing cytopathic effects (Meekins et al. Citation2020).
Based on current understanding, the risk of contracting SARS-CoV-2 during a transplant operation involving pig-derived products should be very low, although it should be extensively investigated. Donor pigs should be tested for viral RNA of SARS-CoV-2 by PCR or antibodies by serology to rule out any infection. This protocol for human blood donors is being used in Wuhan and Hubei provinces, China (Chang et al. Citation2020). Various studies have revealed that different kinds of foods and food products including fruits, vegetables, bread, dairy products, and ready-to-eat foods, meat and meat products may act as potential vehicles/carriers for transmission of SARS-CoV-2 via carry-through or carry-over contaminations, therefore adequate precautious are warranted during handling of foods and in food processing units especially related to meat industry (Faslu Rahman et al. Citation2020a, Citation2020b; Hedman et al. Citation2021; Hu et al. Citation2021; Jo et al. Citation2021; Mallapaty Citation2021; Yekta et al. Citation2021). Hence, conclusive evidence of pigs getting SARS-CoV-2 infection is limited with some studies validating in favor and some against it, therefore explorative studies and larger monitoring investigations are required to conclude towards the role of pigs in SARS-CoV-2 transmisison and spill-over events to humans during the ongoing pandemic (Damas et al. Citation2020; Meekins et al. Citation2020 Zhao et al. Citation2020; Ma and Gong Citation2021; Pickering et al. Citation2021).
7. Conclusion and future prospects
Coronaviruses mostly affect the respiratory and intestinal tracts of both humans and animals. The potential of CoVs to mutate and accommodate to a different new environment renders them a constant threat to humans and animals, particularly in the swine industry workers, veterinarians, and animal handlers. Porcine CoVs, such as TGEV, PRCV, PEDV, PHEV, PDCoV, and SADS-CoV, are more common in piglets than in adults, so a strategic plan is needed to maintain a maternal level of antibody in newborns so that mortality can be reduced. Unified identification methods for TGEV, PRCV, PEDV, PHEV, PDCoV and SADS-CoV, including serological and virological methods, are required to prevent potential outbreaks. Past outbreaks of SARS-CoV and MERS-CoV, along with recent SARS-CoV-2 outbreaks have shown that the virus will cross species barriers and affect people, necessitating close monitoring and prompt updates to avoid potential losses. Since pigs are infected with a variety of CoVs, it is possible that SARS-CoV-2 may also be from pigs, as a result of genetic variation. SARS-CoV-2 infections in animals are becoming more common, highlighting the need to better understand and assess animal vulnerability, which is important for public health and the economy. Because only a few studies have looked into whether pigs can be infected with SARS-CoV, MERS-CoV, and SARS CoV-2, more research is needed to combat future pandemics in pigs and humans.
The recent and rapid global spread of highly pathogenic variants of SADS-CoV, PEDV, and PDCoV highlights the serious One Health threat posed by a newly emerged swine coronavirus, and that highlights the need for resources to better understand the virus and its disease-causing capability in mammals. Researchers must investigate the zoonotic potential of all PCoVs, as well as emerging porcine CoVs like PDCoV and SADS-CoV, so as to avoid future pandemics. Systematic monitoring and tracking of the proliferation of novel variant viruses in human and animal species can be done using the One Health approach. Intensive animal monitoring and risk assessments, particularly during and after an outbreak, will lead to an understanding of the diseases in natural animal reservoirs and to prevent spillover events. Swine farm in rural areas frequently utilize leftover foods from city restaurants as hogwash, without thoroughly fermenting it. Since there is no direct evidence of human-to-swine SARS-CoV-2 transmission, a legal notification must be issued to avoid such a practice, or legislative procedures should be implemented to prevent disease transmission through this route. The rising incidence of novel human and animal CoVs prompts the development of new methods for identifying higher-risk strains that can prevent forthcoming disease outbreaks. The majority of newly emerging human and animal CoVs, including SARS-CoV-2, is thought to have originated in bats and have the ability to adapt and propagate rapidly throughout the world. With increased global travel and frequent human-to-human interaction, it's more important than ever to develop and implement strategies for assessing the potential risk of emerging viruses to One Health globally.
Acknowledgements
All the authors acknowledge and thank their respective Institutes and Universities.
Disclosure statement
All authors declare that there exist no commercial or financial relationships that could, in any way, lead to a potential conflict of interest.
Funding
This compilation is a review article written by its authors and required no substantial funding to be stated. Thank is due to the Faculty of Medicine Siriraj Hospital Mahidol University for paying the publication charge.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- Abdullahi IN, Emeribe AU, Ajayi OA, Oderinde BS, Amadu DO, Osuji AI. 2020. Implications of SARS-CoV-2 genetic diversity and mutations on pathogenicity of the COVID-19 and biomedical interventions. J Taibah Univ Med Sci. 15(4):258–264.
- Ahmed SSSJ, Paramasivam P, Raj K, Kumar V, Murugesan R, Ramakrishnan V. 2020. Regulatory cross talk between SARS-CoV-2 receptor binding and replication machinery in the human host. Front Physiol. 11:802.
- Akimkin V, Beer M, Blome S, Hanke D, Hoper D, Jenckel M, Pohlmann A. 2016. New chimeric porcine coronavirus in swine feces, Germany, 2012. Emerg Infect Dis. 22(7):1314–1315.
- Alexander HK, Day T. 2010. Risk factors for the evolutionary emergence of pathogens. J R Soc Interface. 7(51):1455–1474.
- Alluwaimi AM, Alshubaith IH, Al-Ali AM, Abohelaika S. 2020. The coronaviruses of animals and birds: their zoonosis, vaccines, and models for SARS-CoV and SARS-CoV2. Front Vet Sci. 7:582287.
- Almeida JD, Berry DM, Cunningham CH, Hamre D, Hofstad MS, Mallucci L, McIntosh K, Tyrrell DAJ. 1968. Coronaviruses. Nature 1968; 220(5168): 650
- Alnazawi M, Altaher A, Kandeel M. 2017. Comparative genomic analysis MERS CoV isolated from humans and camels with special reference to virus encoded helicase. Biol Pharm Bull. 40(8):1289–1298.
- Alonso C, Goede DP, Morrison RB, Davies PR, Rovira A, Marthaler DG, Torremorell M. 2014. Evidence of infectivity of airborne porcine epidemic diarrhea virus and detection of airborne viral RNA at long distances from infected herds. Vet Res. 45(1):73.
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med. 26(4):450–452.
- Asrani P, Hasan GM, Sohal SS, Hassan MI. 2020. Molecular basis of pathogenesis of coronaviruses: A comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st century. OMICS. 24(11):634–644.
- Bang FB. 1978. Genetics of resistance of animals to viruses: I. Introduction and studies in mice. Adv Virus Res. 23:269–348.
- Belsham GJ, Rasmussen TB, Normann P, Vaclavek P, Strandbygaard B, Bøtner A. 2016. Characterization of a novel chimeric swine enteric coronavirus from diseased pigs in central eastern Europe in 2016. Transbound Emerg Dis. 63(6):595–601.
- Berger A, Drosten C, Doerr HW, Stürmer M, Preiser W. 2004. Severe acute respiratory syndrome (SARS)-paradigm of an emerging viral infection. J Clin Virol. 29(1):13–22.
- Bevins SN, Lutman M, Pedersen K, Barrett N, Gidlewski T, Deliberto TJ, Franklin AB. 2018. Spillover of swine coronaviruses, United States. Emerg Infect Dis. 24(7):1390–1392.
- Bidokhti MRM, Tråvén M, Krishna NK, Munir M, Belák S, Alenius S, Cortey M. 2013. Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains. J Gen Virol. 94(Pt 9):2036–2049.
- Boley PA, Alhamo MA, Lossie G, Yadav KK, Vasquez-Lee M, Saif LJ, Kenney SP. 2020. Porcine Deltacoronavirus infection and transmission in poultry, United States. Emerg Infect Dis. 26(2):255–265.
- Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, Faccini S, Bonilauri P, Cordioli P, Marthaler D. 2016. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. 22(1):83–87.
- Borremans B, Faust C, Manlove KR, Sokolow SH, Lloyd-Smith JO. 2019. Cross-species pathogen spillover across ecosystem boundaries: mechanisms and theory. Philos Trans R Soc Lond B Biol Sci. 374(1782):20180344.
- Brook CE, Boots M, Chandran K, Dobson AP, Drosten C, Graham AL, Grenfell BT, Müller MA, Ng M, Wang LF, et al. 2020. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 9:e48401. Feb 3
- Castano-Rodriguez C, Honrubia JM, Gutierrez-Alvarez J, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeno JM, Regla-Nava JA, Fernandez-Delgado R, Verdia-Baguena C, Queralt-Martín M, et al. 2018. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 9(3):e02325-17.
- Chang L, Yan Y, Wang L. 2020. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 34(2):75–80.
- Chasey D, Cartwright SF. 1978. Virus-like particles associated with porcine epidemic diarrhoea. Res Vet Sci. 25(2):255–256.
- Chen F, Knutson TP, Rossow S, Saif LJ, Marthaler DG. 2019. Decline of transmissible gastroenteritis virus and its complex evolutionary relationship with porcine respiratory coronavirus in the United States. Sci Rep. 9(1):3953.
- Chen X, Laurent S, Onur OA, Kleineberg NN, Fink GR, Schweitzer F, Warnke C. 2021. A systematic review of neurological symptoms and complications of COVID-19. J Neurol. 268(2):392–402.
- Chen Y, Liu Q, Guo D. 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 92(4):418–423.
- Cheng S, Wu H, Chen Z. 2020. Evolution of transmissible gastroenteritis virus (TGEV): a codon usage perspective. IJMS. 21(21):7898.
- Cheun-Arom T, Temeeyasen G, Tripipat T, Kaewprommal P, Piriyapongsa J, Sukrong S, Chongcharoen W, Tantituvanont A, Nilubol D. 2016. Full-length genome analysis of two genetically distinct variants of porcine epidemic diarrhea virus in Thailand. Infect Genet Evol. 44:114–121.
- Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y, Hu B, Yip CC, Tsang JO, Huang X, et al. 2020. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 1(1):e14–e23.
- Colina SE, Serena MS, Echeverría MG, Metz GE. 2021. Clinical and molecular aspects of veterinary coronaviruses. Virus Res. 297:198382.
- Crawford K, Lager K, Miller L, Opriessnig T, Gerber P, Hesse R. 2015. Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet Res. 46(1):49.
- Cui J, Eden JS, Holmes EC, Wang LF. 2013. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol J. 10:304.
- Cui J, Li F, Shi ZL. 2019. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 17(3):181–192.
- Cui T, Theuns S, Xie J, Broeck W, Nauwynck H. 2020. Role of porcine aminopeptidase N and sialic acids inporcine coronavirus infections in primary porcine enterocytes. Viruses. 12(4):402.
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli KP, Pfenning AR, Zhao H, et al. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA. 117(36):22311–22322.
- de Nova PJG, Cortey M, Díaz I, Puente H, Rubio P, Martín M, Carvajal A. 2020. A retrospective study of porcine epidemic diarrhoea virus (PEDV) reveals the presence of swine enteric coronavirus (SeCoV) since 1993 and the recent introduction of a recombinant PEDV-SeCoV in Spain. Transbound Emerg Dis. 67(6):2911–2922.
- Decaro N, Lorusso A. 2020. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet Microbiol. 244:108693.
- Decaro N, Martella V, Saif LJ, Buonavoglia C. 2020. COVID-19 from veterinary medicine and one health perspectives: what animal coronaviruses have taught us? Res Vet Sci. 131:21–23.
- Dee S, Clement T, Schelkopf A, Nerem J, Knudsen D, Christopher-Hennings J, Nelson E. 2014. An evaluation of contaminated complete feed as a vehicle for porcine epidemic diarrhea virus infection of naïve pigs following consumption via natural feeding behavior: proof of concept. BMC Vet Res. 10:176.
- Dee S, Neill C, Singrey A, Clement T, Cochrane R, Jones C, Patterson G, Spronk G, Christopher-Hennings J, Nelson E. 2016. Modeling the transboundary risk of feed ingredients contaminated with porcine epidemic diarrhea virus. BMC Vet Res. 12:51.
- Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, Singh KP, Chaicumpa W, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. Coronavirus disease 2019-COVID-19. Clin Microbiol Rev. 33(4):e00028-20.
- Dhama K, Pawaiya RVS, Chakrabort S, Tiwari R, Saminathan M, Verma AK. 2014a. Coronavirus infection in equines: a review. Asian J Anim Vet Adv. 9(3):164–176.
- Dhama K, Rajagunalan S, Chakraborty S, Verma AK, Kumar A, Tiwari R, Kapoor S. 2013a. Food-borne pathogens of animal origin-diagnosis, prevention, control and their zoonotic significance: a review. Pak J Biol Sci. 16(20):1076–1085.
- Dhama K, Singh SD, Barathidasan R, Desingu PA, Chakraborty S, Tiwari R, Kumar MA. 2014b. Emergence of Avian Infectious Bronchitis Virus and its variants need better diagnosis, prevention and control strategies: a global perspective. Pak J Biol Sci. 17(6):751–767.
- Dhama K, Chakraborty S, Kapoor S, Tiwari R, Kumar A, Deb R, Rajagunalan S, Singh R, Vora K, Natesan S. 2013b. One world, one health - veterinary perspectives. Adv Anim Vet Sci. 1(1):5–13.
- Dhama K, Verma AK, Tiwari R, Chakraborty S, Vora K, Kapoor S, Deb R, Karthik K, Singh R, Munir M, et al. 2013c. A perspective on applications of geographical information system (GIS): an advanced tracking tool for disease surveillance and monitoring in veterinary epidemiology. Adv Anim Vet Sci. 1(1):14–24.
- Diep NV, Sueyoshi M, Izzati U, Fuke N, Teh APP, Lan NT, Yamaguchi R. 2018. Appearance of US-like porcine epidemic diarrhoea virus (PEDV) strains before US outbreaks and genetic heterogeneity of PEDVs collected in Northern Vietnam during 2012-2015. Transbound Emerg Dis. 65(1):e83–e93.
- Ding G, Fu Y, Li B, Chen J, Wang J, Yin B, Sha W, Liu G. 2020. Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China. Transbound Emerg Dis. 67(2):678–685.
- Domingo E, Perales C. 2019. Viral quasispecies. PLoS Genet. 15(10):e1008271.
- Dong B, Gao W, Lu H, Zhao K, Ding N, Liu W, Zhao J, Lan Y, Tang B, Jin Z, et al. 2015. A small region of porcine hemagglutinating encephalomyelitis virus spike protein interacts with the neural cell adhesion molecule. Intervirology. 58(2):130–137.
- Dong B, Lu H, Zhao K, Liu W, Gao W, Lan Y, Zhao J, Tang B, Song D, He W, et al. 2014. Identification and genetic characterization of porcine hemagglutinating encephalomyelitis virus from domestic piglets in China. Arch Virol. 159(9):2329–2337.
- Dong BQ, Liu W, Fan XH, Vijaykrishna D, Tang XC, Gao F, Li LF, Li GJ, Zhang JX, Yang LQ, et al. 2007. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J Virol. 81(13):6920–6926.
- Doyle LP, Hutchings LM. 1946. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc. 108:257–259.
- Du Toit A. 2020. Coronavirus replication factories. Nat Rev Microbiol. 18(8):411.
- Duffy S. 2018. Why are RNA virus mutation rates so damn high? PLoS Biol. 16(8):e3000003.
- Edwards CE, Yount BL, Graham RL, Leist SR, Hou YJ, Dinnon KH, Sims AC, Swanstrom J, Gully K, Scobey TD, et al. 2020. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc Natl Acad Sci USA. 117(43):26915–26925.
- El-Tholoth M, Bai H, Mauk MG, Saif L, Bau HH. 2021. A portable, 3D printed, microfluidic device for multiplexed, real time, molecular detection of the porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine deltacoronavirus at the point of need. Lab Chip. 21(6):1118–1130.
- Enjuanes L, van der Zeijst BAM. 1995. Molecular basis of transmissible gastroenteritis virus epidemiology. Coronaviridae. 337–376.
- Fang P, Fang L, Hong Y, Liu X, Dong N, Ma P, Bi J, Wang D, Xiao S. 2017. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J Gen Virol. 98(2):173–178.
- Faslu Rahman CK, Sharun K, Jose B, Dhama K. 2020a. COVID-19 and food safety: implications and opportunities to improve the food supply chain. J Exp Biol and Agri Sci. 8:S34–S41.
- Faslu Rahman CK, Sharun K, Tiwari R, Bilal M, Dhama K. 2020b. Can unconventional meat or bushmeat act as a source for SARS-COV-2. JEBAS. 8(6):709–720.
- Fehr AR, Perlman S. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 1282:1–23.
- Fenner F. 2017. Chapter 24. Coronaviridae. In Maclachlan NJ, Dubovi EJ, editors. Fenner’s veterinary virology. 5th ed. New York: Academic Press.
- Forni D, Cagliani R, Clerici M, Sironi M. 2017. Molecular evolution of human coronavirus genomes. Trends Microbiol. 25(1):35–48.
- Frutos R, Serra-Cobo J, Pinault L, Lopez Roig M, Devaux CA. 2021. Emergence of bat-related betacoronaviruses: hazard and risks. Front Microbiol. 12:591535.
- Fumagalli M, Pozzoli U, Cagliani R, Comi GP, Bresolin N, Clerici M, Sironi M. 2010. Genome-wide identification of susceptibility alleles for viral infections through a population genetics approach. PLoS Genet. 6(2):e1000849.
- Gao W, Zhao K, Zhao C, Du C, Ren W, Song D, Lu H, Chen K, Li Z, Lan Y, et al. 2011. Vomiting and wasting disease associated with hemagglutinating encephalomyelitis viruses infection in piglets in Jilin, China. Virol J. 8:130.
- Garcia GG, Aquino MAD, Balbin MM, Belotindos LP, Supnet JG, Mingala CN. 2018. Characterisation of porcine epidemic diarrhea virus isolates during the 2014-2015 outbreak in the Philippines. Virusdisease. 29(3):342–348.
- Ghosh S, Malik YS. 2020. Drawing comparisons between SARS-CoV-2 and the animal coronaviruses. Microorganisms. 8(11):1840.
- Goldstein SA, Brown J, Pedersen BS, Quinlan AR, Elde NC. 2021. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. bioRxiv [Preprint]429646.
- Gong L, Li J, Zhou Q, Xu Z, Chen L, Zhang Y, Xue C, Wen Z, Cao Y. 2017. A new bat-HKU2-like coronavirus in swine, China, 2017. Emerg Infect Dis. 23(9):1607–1609.
- Greig AS, Mitchell D, Corner AH, Bannister GL, Meads EB, Julian RJ. 1962. A hemagglutinating virus producing encephalomyelitis in baby pigs. Can J Comp Med Vet Sci. 26(3):49–56.
- Guo R, Fan B, Chang X, Zhou J, Zhao Y, Shi D, Yu Z, He K, Li B. 2020. Characterization and evaluation of the pathogenicity of a natural recombinant transmissible gastroenteritis virus in China. Virology. 545:24–32.
- Haake C, Cook S, Pusterla N, Murphy B. 2020. Coronavirus infections in companion animals: virology, epidemiology, clinical and pathologic features. Viruses. 12(9):1023.
- He WT, Ji X, He W, Dellicour S, Wang S, Li G, Zhang L, Gilbert M, Zhu H, Xing G, et al. 2020. Genomic epidemiology, evolution, and transmission dynamics of porcine Deltacoronavirus. Mol Biol Evol. 37(9):2641–2654.
- Hedman HD, Krawczyk E, Helmy YA, Zhang L, Varga C. 2021. Host diversity and potential transmission pathways of SARS-CoV-2 at the human-animal interface. Pathogens. 10(2):180.
- Heegaard PMH, Sturek M, Alloosh M, Belsham GJ. 2020. Animal models for COVID-19: more to the picture than ACE2, rodents, ferrets, and non-human primates. A case for porcine respiratory coronavirus and the obese Ossabaw pig. Front Microbiol. 11:573756.
- Hu H, Jung K, Vlasova AN, Saif LJ. 2016. Experimental infection of gnotobiotic pigs with the cell-culture-adapted porcine Deltacoronavirus strain OH-FD22. Arch Virol. 161(12):3421–3434.
- Hu L, Gao J, Yao L, Zeng L, Liu Q, Zhou Q, Zhang H, Lu D, Fu J, Liu QS, et al. 2021. Evidence of foodborne transmission of the coronavirus (COVID-19) through the animal products food supply chain. Environ Sci Technol. 55(5):2713–2716.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395(10223):497–506.
- Iseki H, Watanabe S, Mase M. 2021. A potential system for the isolation and propagation of porcine deltacoronavirus using embryonated chicken eggs. J Virol Methods. 290:114068.
- Ismail MM, Cho KO, Ward LA, Saif LJ, Saif YM. 2001. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. 45(1):157–163.
- Iturriza-Gomara M, O'Brien SJ. 2016. Foodborne viral infections. Curr Opin Infect Dis. 29(5):495–501.
- Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S, Nonthabenjawan N, Kesdaengsakonwut S, Amonsin A. 2016. Porcine Deltacoronavirus, Thailand, 2015. Emerg Infect Dis. 22(4):757–759.
- Jelinek HF, Mousa M, Alefishat E, Osman W, Spence I, Bu D, Feng SF, Byrd J, Magni PA, Sahibzada S, et al. 2021. Evolution, ecology, and zoonotic transmission of betacoronaviruses: a review. Front Vet Sci. 8:644414.
- Ji W, Wang W, Zhao X, Zai J, Li X. 2020. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 92(4):433–440.
- Jo WK, de Oliveira-Filho EF, Rasche A, Greenwood AD, Osterrieder K, Drexler JF. 2021. Potential zoonotic sources of SARS-CoV-2 infections. Transbound Emerg Dis. 68(4):1824–1834.
- Jung K, Hu H, Saif LJ. 2016. Porcine Deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 226:50–59.
- Jung K, Saif LJ, Wang Q. 2020. Porcine epidemic diarrhea virus (PEDV): an update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 286:198045.
- Kahn JS, McIntosh K. 2005. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 24(11 Suppl):S223–S227.
- Kautz TF, Forrester NL. 2018. RNA virus fidelity mutants: a useful tool for evolutionary biology or a complex challenge? Viruses. 10(11):600.
- Kenney SP, Wang Q, Vlasova A, Jung K, Saif L. 2021. Naturally occurring animal coronaviruses as models for studying highly pathogenic human coronaviral disease. Vet Pathol. 58(3):438–452.
- Khamassi Khbou M, Daaloul Jedidi M, Bouaicha Zaafouri F, Benzarti M. 2021. Coronaviruses in farm animals: epidemiology and public health implications. Vet Med Sci. 7(2):322–347.
- Kim L, Chang KO, Sestak K, Parwani A, Saif LJ. 2000. Development of a reverse transcription-nested polymerase chain reaction assay for differential diagnosis of transmissible gastroenteritis virus and porcine respiratory coronavirus from feces and nasal swabs of infected pigs. J Vet Diagn Invest. 12(4):385–388.
- Koonpaew S, Teeravechyan S, Frantz PN, Chailangkarn T, Jongkaewwattana A. 2019. PEDV and PDCoV pathogenesis: the interplay between host innate immune responses and porcine enteric coronaviruses. Front Vet Sci. 6:34.
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, et al. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 182(4):812–827.e19.
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 348(20):1953–1966.
- Kuo L, Hurst-Hess KR, Koetzner CA, Masters PS. 2016. Analyses of coronavirus assembly interactions with interspecies membrane and nucleocapsid protein chimeras. J Virol. 90(9):4357–4368.
- Lau SKP, Wong EYM, Tsang CC, Ahmed SS, Au-Yeung RKH, Yuen KY, Wernery U, Woo PCY. 2018. Discovery and sequence analysis of four deltacoronaviruses from birds in the Middle East reveal interspecies jumping with recombination as a potential mechanism for avian-to-avian and avian-to-mammalian transmission. J Virol. 92(15):e00265-18.
- Lawhorn B. 1999. Diarrheal disease in show swine. Texas Agricultural Extension Service, United States: Texas A & M University System.
- Le VP, Song S, An BH, Park GN, Pham NT, Le DQ, Nguyen VT, Vu TTH, Kim KS, Choe S, et al. 2018. A novel strain of porcine deltacoronavirus in Vietnam. Arch Virol. 163(1):203–207. Epub 2017 Oct 11.
- Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, Bonny TS, Loeb JC, Telisma T, Chavannes S, et al. 2021a. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv.
- Lednicky JA, Tagliamonte MS, White SK, Elbadry MA, Alam MM, Stephenson CJ, Bonny TS, Loeb JC, Telisma T, Chavannes S, et al. 2021b. Independent infections of porcine deltacoronavirus among Haitian children. Nature. 600(7887):133–137.
- Lee JH, Chung HC, Nguyen VG, Moon HJ, Kim HK, Park SJ, Lee CH, Lee GE, Park BK. 2016. Detection and phylogenetic analysis of porcine Deltacoronavirus in Korean Swine Farms, 2015. Transbound Emerg Dis. 63(3):248–252.
- Lee S, Lee C. 2014. Complete genome characterization of Korean porcine Deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. 2(6):e01191-14.
- Lee YJ, Lee C. 2018. Porcine deltacoronavirus induces caspase-dependent apoptosis through activation of the cytochrome c-mediated intrinsic mitochondrial pathway. Virus Res. 253:112–123.
- Li D, Feng H, Liu Y, Chen Y, Wei Q, Wang J, Liu D, Huang H, Su Y, Wang D, et al. 2018a. Molecular evolution of porcine epidemic diarrhea virus and porcine deltacoronavirus strains in Central China. Res Vet Sci. 120:63–69.
- Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J. 2014. Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announc. 2(2):e00278-14.
- Li J, Yuan C, Liu P, Li Y, Zhang P, Yang Q. 2021. Red blood cells serve as a vehicle for PEDV transmission. Vet Microbiol. 257:109081.
- Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. 2018b. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. 115(22):E5135–E5143.
- Li Y, Wu Q, Huang L, Yuan C, Wang J, Yang Q. 2018c. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat Commun. 9(1):3811.
- Li Z, He W, Lan Y, Zhao K, Lv X, Lu H, Ding N, Zhang J, Shi J, Shan C, et al. 2016. The evidence of porcine hemagglutinating encephalomyelitis virus induced nonsuppurative encephalitis as the cause of death in piglets. PeerJ. 4:e2443.
- Liu C, Tang J, Ma Y, Liang X, Yang Y, Peng G, Qi Q, Jiang S, Li J, Du L, et al. 2015. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol. 89(11):6121–6125.
- Liu C, Zhang X, Zhang Z, Chen R, Zhang Z, Xue Q. 2017. Complete genome characterization of novel Chinese porcine Deltacoronavirus strain SD. Genome Announc. 5(40):e00930-17.
- Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM. 2014. The evolution and genetics of virus host shifts. PLoS Pathog. 10(11):e1004395.
- Lorbach JN, Wang L, Nolting JM, Benjamin MG, Killian ML, Zhang Y, Bowman AS. 2017. Porcine hemagglutinating encephalomyelitis virus and respiratory disease in exhibition swine, Michigan, USA, 2015. Emerg Infect Dis. 23(7):1168–1171.
- Lorsirigool A, Saeng-Chuto K, Temeeyasen G, Madapong A, Tripipat T, Wegner M, Tuntituvanont A, Intrakamhaeng M, Nilubol D. 2016. The first detection and full-length genome sequence of porcine deltacoronavirus isolated in Lao PDR. Arch Virol. 161(10):2909–2911.
- Lorsirigool A, Saeng-Chuto K, Madapong A, Temeeyasen G, Tripipat T, Kaewprommal P, Tantituvanont A, Piriyapongsa J, Nilubol D. 2017. The genetic diversity and complete genome analysis of two novel porcine deltacoronavirus isolates in Thailand in 2015. Virus Genes. 53(2):240–248.
- Lu G, Wang Q, Gao GF. 2015. Bat-to-human: spike features determining 'host jump' of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 23(8):468–478.
- Luo SX, Fan JH, Opriessnig T, Di JM, Liu BJ, Zuo YZ. 2017. Development and application of a recombinant M protein-based indirect ELISA for the detection of porcine deltacoronavirus IgG antibodies. J Virol Methods. 249:76–78.
- Luo X, Zhou GZ, Zhang Y, Peng LH, Zou LP, Yang YS. 2020. Coronaviruses and gastrointestinal diseases. Mil Med Res. 7(1):49.
- Ma C, Gong C. 2021. ACE2 models of frequently contacted animals provide clues of their SARS-CoV-2 S protein affinity and viral susceptibility. J Med Virol. 93(7):4469–4479.
- Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, Li J. 2015. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio. 6(2):e00064. Mar 10
- Ma Y, Zhang Y, Liang X, Oglesbee M, Krakowka S, Niehaus A, Wang G, Jia A, Song H, Li J. 2016. Two-way antigenic cross-reactivity between porcine epidemic diarrhea virus and porcine Deltacoronavirus. Vet Microbiol. 186:90–96.
- Madapong A, Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Srijangwad A, Tripipat T, Wegner M, Nilubol D. 2016. Complete genome sequence of porcine deltacoronavirus isolated in Thailand in 2015. Genome Announc. 4(3):e00408-16.
- Magtoto R, Poonsuk K, Baum D, Zhang J, Chen Q, Ji J, Piñeyro P, Zimmerman J, Giménez-Lirola LG. 2019. Evaluation of the serologic cross-reactivity between transmissible gastroenteritis coronavirus and porcine respiratory coronavirus using commercial blocking enzyme-linked immunosorbent assay kits. mSphere. 4(2):e00017-19.
- Malik YS, Sircar S, Bhat S, Sharun K, Dhama K, Dadar M, Tiwari R, Chaicumpa W. 2020. Emerging novel coronavirus (2019-nCoV)-current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 40(1):68–76.
- Mallapaty S. 2021. The search for animals harbouring coronavirus - and why it matters. Nature. 591(7848):26–28.
- Mandary MB, Masomian M, Poh CL. 2019. Impact of RNA virus evolution on quasispecies formation and virulence. Int J Mol Sci. 20(18):4657.
- Mandelik R, Sarvas M, Jackova A, Salamunova S, Novotny J, Vilcek S. 2018. First outbreak with chimeric swine enteric coronavirus (SeCoV) on pig farms in Slovakia - lessons to learn. Acta Vet Hung. 66(3):488–492.
- Marthaler D, Jiang Y, Collins J, Rossow K. 2014. Complete genome sequence of strain SDCV/USA/Illinois121/2014, a porcine Deltacoronavirus from the United States. Genome Announc. 2(2):e00218-14.
- Meekins DA, Morozov I, Trujillo JD, Gaudreault NN, Bold D, Carossino M, Artiaga BL, Indran SV, Kwon T, Balaraman V, et al. 2020. Susceptibility of swine cells and domestic pigs to SARS-CoV-2. Emerg Microbes Infect. 9(1):2278–2288.
- Miłek J, Blicharz-Domańska K. 2018. Coronaviruses in avian species - review with focus on epidemiology and diagnosis in wild birds. J Vet Res. 62(3):249–255.
- Miller CS. 2005. Pleiotropic mechanisms of virus survival and persistence. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 100(2 Suppl):S27–S36.
- Moon HW. 1978. Mechanisms in the pathogenesis of diarrhea: a review. J Am Vet Med Assoc. 172(4):443–448.
- Mora-Diaz JC, Magtoto R, Houston E, Baum D, Carrillo-Ávila JA, Temeeyasen G, Zimmerman J, Piñeyro P, Giménez-Lirola, L, 2020. Detecting and monitoring porcine hemagglutinating encephalomyelitis virus, an under-researched Betacoronavirus. mSphere. 5(3):e00199-20.
- Mora-Diaz JC, Piñeyro PE, Houston E, Zimmerman J, Giménez-Lirola LG. 2019. Porcine hemagglutinating encephalomyelitis virus: a review. Front Vet Sci. 6:53.
- Mora-Diaz JC, Piñeyro PE, Rauh R, Nelson W, Sankoh Z, Gregg E, Carrillo-Ávila JA, Shen H, Nelli RK, Zimmerman JJ, et al. 2021. Porcine hemagglutinating encephalomyelitis virus infection in vivo and ex vivo. J Virol. 95(12):e02335-20.
- Mostl K. 1990. Coronaviridae, pathogenetic and clinical aspects: an update. Comp Immunol Microbiol Infect Dis. 13(4):169–180.
- Nasir A, Sun FJ, Kim KM, Caetano-Anollés G. 2015. Untangling the origin of viruses and their impact on cellular evolution. Ann N Y Acad Sci. 1341:61–74.
- Niederwerder MC, Hesse RA. 2018. Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound Emerg Dis. 65(3):660–675.
- Oberemok VV, Laikova KV, Yurchenko KA, Fomochkina II, Kubyshkin AV. 2020. SARS-CoV-2 will continue to circulate in the human population: an opinion from the point of view of the virus-host relationship. Inflamm Res. 69(7):635–640.
- Ojkic D, Hazlett M, Fairles J, Marom A, Slavic D, Maxie G, Alexandersen S, Pasick J, Alsop J, Burlatschenko S. 2015. The first case of porcine epidemic diarrhea in Canada. Can Vet J. 56(2):149–152.
- Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature. 546(7660):646–650.[28636590]
- Opriessnig T, Huang YW. 2020. Coronavirus disease 2019 (COVID-19) outbreak: could pigs be vectors for human infections? Xenotransplantation. 27(2):e12591.
- Pan Y, Tian X, Qin P, Wang B, Zhao P, Yang YL, Wang L, Wang D, Song Y, Zhang X, et al. 2017. Discovery of a novel swine enteric Alphacoronavirus (SeACoV) in southern China. Vet Microbiol. 211:15–21.
- Parkhe P, Verma S. 2021. Evolution, interspecies transmission, and zoonotic significance of animal coronaviruses. Front Vet Sci. 8:719834.
- Peng JY, Punyadarsaniya D, Shin DL, Pavasutthipaisit S, Beineke A, Li G, Wu NH, Herrler G. 2020. The cell tropism of porcine respiratory coronavirus for airway epithelial cells is determined by the expression of porcine aminopeptidase N. Viruses. 12(11):1211. Oct 23
- Pensaert M, Callebaut P, Vergote J. 1986. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q. 8(3):257–261.
- Pensaert MB, Martelli P. 2016. Porcine epidemic diarrhea: a retrospect from Europe and matters of debate. Virus Res. 226:1–6.
- Perez-Rivera C, Ramirez-Mendoza H, Mendoza-Elvira S, Segura-Velazquez R, Sanchez-Betancourt JI. 2019. First report and phylogenetic analysis of porcine deltacoronavirus in Mexico. Transbound Emerg Dis. 66(4):1436–1441.
- Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, Nicholls J, Yee WK, Yan WW, Cheung MT, Cheng VC, Chan KH, Tsang DN, Yung RW, Ng T K, Yuen KY. 2003. SARS study group. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 361(9366):1319–1325.
- Pickering BS, Smith G, Pinette MM, Embury-Hyatt C, Moffat E, Marszal P, Lewis CE. 2021. Susceptibility of domestic swine to experimental infection with severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 27(1):104–112.
- Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, Lloyd-Smith JO. 2017. Pathways to zoonotic spillover. Nat Rev Microbiol. 15(8):502–510.
- Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, Dhama K, Yatoo MI, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 28(2):174–184.
- Rabaan AA, Al-Ahmed SH, Sah R, Alqumber MA, Haque S, Patel SK, Pathak M, Tiwari R, Yatoo MI, Haq AU, et al. 2021. MERS-CoV: epidemiology, molecular dynamics, therapeutics, and future challenges. Ann Clin Microbiol Antimicrob. 20(1):8.
- Ren X, Glende J, Al-Falah M, de Vries V, Schwegmann-Wessels C, Qu X, Tan L, Tschernig T, Deng H, Naim HY, et al. 2006. Analysis of ACE2 in polarized epithelial cells: surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J Gen Virol. 87(Pt 6):1691–1695.
- Reveles-Felix S, Carreon-Napoles R, Mendoza-Elvira S, Quintero-Ramírez V, Garcia-Sanchez J, Martínez-Bautista R, Saavedra-Montaiez M, Mosqueda Gualito JJ, Sanchez-Betancourt JI. 2020. Emerging strains of porcine epidemic diarrhoea virus (PEDv) in Mexico. Transbound Emerg Dis. 67(2):1035–1041.
- Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, de Meulder D, van Amerongen G, van den Brand J, Okba NMA, et al. 2020. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 368(6494):1012–1015.
- Roe CK, Alexander TJ. 1958. A disease of nursing pigs previously unreported in Ontario. Can J Comp Med Vet Sci. 22(9):305–307.
- Saade G, Deblanc C, Bougon J, Marois-Créhan C, Fablet C, Auray G, Belloc C, Leblanc-Maridor M, Gagnon CA, Zhu J, et al. 2020. Coinfections and their molecular consequences in the porcine respiratory tract. Vet Res. 51(1):80.
- Saeng-Chuto K, Lorsirigool A, Temeeyasen G, Vui DT, Stott CJ, Madapong A, Tripipat T, Wegner M, Intrakamhaeng M, Chongcharoen W, et al. 2017. Different lineage of porcine Deltacoronavirus in Thailand, Vietnam and Lao PDR in 2015. Transbound Emerg Dis. 64(1):3–10.
- Saif LJ, Jung K. 2020. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J Clin Microbiol. 58(8):e01355-20.
- Saif LJ, Sestak K. 2006. Transmissible gastroenteritis and porcine respiratory coronavirus. In Straw B, Zimmerman JJ, D’Allaire S, Taylor DJ, editors. Diseases of swine. 9th ed. Ames (IA): Blackwell Publishing; p. 489.
- Saif LJ, Wang Q, Vlasova AN, Jung K, Xiao S. 2019. Coronaviruses. In Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Zhang J, editors. Diseases of swine. Hoboken (NJ): John Wiley and Sons, Inc; p. 488–529.
- Salata C, Calistri A, Parolin C, Palù G. 2019. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathog Dis. 77(9):1ftaa006.
- Sasseville AM, Boutin M, Gélinas AM, Dea S. 2002. Sequence of the 3'-terminal end (8.1 kb) of the genome of porcine haemagglutinating encephalomyelitis virus: comparison with other haemagglutinating coronaviruses. J Gen Virol. 83(Pt 10):2411–2416.
- Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, et al. 2020. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. 1(5):e218–e225.
- Schumacher LL, Woodworth JC, Jones CK, Chen Q, Zhang J, Gauger PC, Stark CR, Main RG, Hesse RA, Tokach MD, et al. 2016. Evaluation of the minimum infectious dose of porcine epidemic diarrhea virus in virus-inoculated feed. Am J Vet Res. 77(10):1108–1113.
- Shaw PD, Patel N, Patil S, Samuel R, Khanna P, Prajapati B, Sharun K, Tiwari R, Dhama K, Natesan S. 2020. Comparative evaluation of the origin, evolution, transmission, diagnosis, and vaccine development of three highly pathogenic human coronaviruses (SARS-COV, MERS-COV and SARS-COV-2). J Exp Biol and Agri Sci. 8(Spl-1-SARS-CoV-2):S103–S113.
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, et al. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 368(6494):1016–1020.
- Sinha A, Gauger P, Zhang J, Yoon KJ, Harmon K. 2015. PCR-based retrospective evaluation of diagnostic samples for emergence of porcine Deltacoronavirus in US swine. Vet Microbiol. 179(3-4):296–298.
- Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, et al. 2015. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 62(6):575–580.
- Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, Zhu H, Zhao W, Han Y, Qin C. 2019. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 11(1):59.
- Steinrigl A, Fernández SR, Stoiber F, Pikalo J, Sattler T, Schmoll F. 2015. First detection, clinical presentation and phylogenetic characterization of porcine epidemic diarrhea virus in Austria. BMC Vet Res. 11:310.
- Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, et al. 2013. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. 25(5):649–654.
- Stott CJ, Sawattrakool K, Saeng-Chuto K, Tantituvanont A, Nilubol D. 2021. The phylodynamics of emerging porcine deltacoronavirus (PDCoV) in Southeast Asia. Transbound Emerg Dis
- Streicker DG, Turmelle AS, Vonhof MJ, Kuzmin IV, McCracken GF, Rupprecht CE. 2010. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science. 329(5992):676–679.
- Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao, GF 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24(6):490–502.
- Tan L, Li Y, He J, Hu Y, Cai X, Liu W, Liu T, Wang J, Li Z, Yuan X, Zhan Y, Yang L, Deng Z, Wang N, Yang Y, Wang A. 2020. Epidemic and genetic characterization of porcine epidemic diarrhea virus strains circulating in the regions around Hunan, China, during 2017-2018. Arch Virol. 165(4):877–889
- Teeravechyan S, Frantz PN, Wongthida P, Chailangkarn T, Jaru-Ampornpan P, Koonpaew S, Jongkaewwattana A. 2016. Deciphering the biology of porcine epidemic diarrhea virus in the era of reverse genetics. Virus Res. 226:152–171.
- Theuns S, Conceicao-Neto N, Christiaens I, Zeller M, Desmarets LM, Roukaerts ID, Acar DD, Heylen E, Matthijnssens J, Nauwynck HJ. 2015. Complete genome sequence of a porcine epidemic diarrhea virus from a novel outbreak in Belgium, January 2015. Genome Announc. 3(3):e00506-15.
- Tiwari R, Dhama K, Sharun K, Iqbal Yatoo M, Malik YS, Singh R, Michalak I, Sah R, Bonilla-Aldana DK, Rodriguez-Morales AJ. 2020. COVID-19: animals, veterinary and zoonotic links. Vet Q. 40(1):169–182.
- Trovato M, Sartorius R, D'Apice L, Manco R, De Berardinis P. 2020. Viral emerging diseases: challenges in developing vaccination strategies. Front Immunol. 11:2130.
- Trujillo-Ortega ME, Beltrán-Figueroa R, García-Hernández ME, Juárez-Ramírez M, Sotomayor-González A, Hernández-Villegas EN, Becerra-Hernández JF, Sarmiento-Silva RE. 2016. Isolation and characterization of porcine epidemic diarrhea virus associated with the 2014 disease outbreak in Mexico: case report. BMC Vet Res. 12(1):132.
- Turlewicz-Podbielska H, Pomorska-Mól M. 2021. Porcine coronaviruses: overview of the state of the art. Virol Sin. 15:1–19.
- Valko A, Bálint Á, Bozsa Á, Cságola A. 2019. Prevalence of antibodies against transmissible gastroenteritis virus (TGEV) in Hungary. Vet Anim Sci. 7:100042.
- Van Sluijs L, Pijlman GP, Kammenga JE. 2017. Why do individuals differ in viral susceptibility? A story told by model organisms. Viruses. 9(10):284.
- V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 19(3):155–170.
- Vlasova AN, Kenney SP, Jung K, Wang Q, Saif LJ. 2021. Deltacoronavirus evolution and transmission: current scenario and evolutionary perspectives. Front Vet Sci. 7:626785.
- Vlasova AN, Wang Q, Jung K, Langel SN, Malik YS, Saif LJ. 2020. Porcine coronaviruses. Emerg Transbound Animal Viruses. 23:79–110.
- Vlok M, Lang AS, Suttle CA. 2019. Marine RNA virus quasispecies are distributed throughout the oceans. mSphere. 4(2):e00157-19.
- Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decadelong structural studies of SARS coronavirus. J Virol. 94(7):e00127-20.
- Wang B, Liu Y, Ji CM, Yang YL, Liang QZ, Zhao P, Xu LD, Lei XM, Luo WT, Qin P, et al. 2018. Porcine Deltacoronavirus engages the transmissible gastroenteritis virus functional receptor porcine aminopeptidase N for infectious cellular entry. J Virol. 92(12):e00318-18.
- Wang J, Li Y, Wang S, Liu F. 2020a. Dynamics of transmissible gastroenteritis virus internalization unraveled by single-virus tracking in live cells . Faseb J. 34(3):4653–4669.
- Wang L, Byrum B, Zhang Y. 2014. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. 20(7):1227–1230.
- Wang Q, Vlasova AN, Kenney SP, Saif LJ. 2019. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. 34:39–49.
- Wang Y, Liu M, Gao J. 2020c. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci USA. 117(25):13967–13974.
- Wei G, Luo S, Wu W, Hu J, Zhou R. 2021. Activation of interleukin-1β release and pyroptosis by transmissible gastroenteritis virus is dependent on the NOD-like receptor protein 3 inflammasome in porcine intestinal epithelial cell line. Viral Immunol.
- Weingartl HM, Copps J, Drebot MA, Marszal P, Smith G, Gren J, Andova M, Pasick J, Kitching P, Czub M. 2004. Susceptibility of pigs and chickens to SARS coronavirus. Emerg Infect Dis. 10(2):179–184.
- Weng L, Weersink A, Poljak Z, de Lange K, von Massow M. 2016. An economic evaluation of intervention strategies for porcine epidemic diarrhea (PED). Prev Vet Med. 134:58–68.
- White JM, Whittaker GR. 2016. Fusion of enveloped viruses in endosomes. Traffic. 17(6):593–614.
- Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, et al. 2012. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J Virol. 86(7):3995–4008.
- Wu HY, Guy JS, Yoo D, Vlasak R, Urbach E, Brian DA. 2003. Common RNA replication signals exist among group 2 coronaviruses: evidence for in vivo recombination between animal and human coronavirus molecules. Virology. 315(1):174–183.
- Wu L, Kewal Ramani VN. 2006. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat Rev Immunol. 6(11):859–868.
- Wu Y, Zhang H, Shi Z, Chen J, Li M, Shi H, Shi D, Guo L, Feng L. 2020. Porcine epidemic diarrhea virus nsp15 antagonizes interferon signalling by RNA degradation of TBK1 and IRF3. Viruses. 12(6):599.
- Xia L, Yang Y, Wang J, Jing Y, Yang Q. 2018. Impact of TGEV infection on the pig small intestine. Virol J. 15(1):102.
- Xu Z, Zhang Y, Gong L, Huang L, Lin Y, Qin J, Du Y, Zhou Q, Xue C, Cao Y. 2019. Isolation and characterization of a highly pathogenic strain of porcine enteric Alphacoronavirus causing watery diarrhoea and high mortality in newborn piglets. Transbound Emerg Dis. 66(1):119–130.
- Yaeger M, Funk N, Hoffman L. 2002. A survey of agents associated with neonatal diarrhea in Iowa swine including Clostridium difficile and porcine reproductive and respiratory syndrome virus. J Vet Diagn Invest. 14(4):281–287.
- Yang YL, Liang QZ, Xu SY, Mazing E, Xu GH, Peng L, Qin P, Wang B, Huang YW. 2019. Characterization of a novel bat-HKU2-like swine enteric alphacoronavirus (SeACoV) infection in cultured cells and development of a SeACoV infectious clone. Virology. 536:110–118.
- Ye X, Chen Y, Zhu X, Guo J, Xie D, Hou Z, Xu S, Zhou J, Fang L, Wang D, et al. 2020. Cross-species transmission of Deltacoronavirus and the origin of porcine Deltacoronavirus. Evol Appl. 13(9):2246–2253.
- Yekta R, Vahid-Dastjerdi L, Norouzbeigi S, Mortazavian AM. 2021. Food products as potential carriers of SARS-CoV-2. Food Control. 123:107754.
- Younes S, Younes N, Shurrab F, Nasrallah GK. 2021. Severe acute respiratory syndrome coronavirus-2 natural animal reservoirs and experimental models: systematic review. Rev Med Virol. 31(4):e2196.
- Yuan D, Yan Z, Li M, Wang Y, Su M, Sun D. 2021. Isolation and characterization of a porcine transmissible gastroenteritis coronavirus in Northeast China. Front Vet Sci. 8:611721.
- Zhang G, Li B, Yoo D, Qin T, Zhang X, Jia Y, Cui S. 2021a. Animal coronaviruses and SARS-CoV-2. Transbound Emerg Dis. 68(3):1097–1110.
- Zhang H, Liang Q, Li B, Cui X, Wei X, Ding Q, Wang Y, Hu H. 2019. Prevalence, phylogenetic and evolutionary analysis of porcine Deltacoronavirus in Henan province, China. Prev Vet Med. 166:8–15. doi: 10.1016/j.prevetmed.2019.02.017.
- Zhang HL, Li YM, Sun J, Zhang YY, Wang TY, Sun MX, Wang MH, Yang YL, Hu XL, Tang YD, et al. 2021c. Evaluating angiotensin-converting enzyme 2-mediated SARS-CoV-2 entry across species. J Biol Chem. 296:100435.
- Zhang J. 2016. Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 226:71–84.
- Zhang X, Zhu Y, Zhu X, Shi H, Chen J, Shi D, Yuan J, Cao L, Liu J, Dong H, et al. 2017. Identification of a natural recombinant transmissible gastroenteritis virus between Purdue and Miller clusters in China. Emerg Microbes Infect. 6(8):e74.
- Zhao J, Cui W, Tian BP. 2020. The potential intermediate hosts for SARS-CoV-2. Front Microbiol. 11:580137.
- Zhou L, Li QN, Su JN, Chen GH, Wu ZX, Luo Y, Wu RT, Sun Y, Lan T, Ma JY. 2019. The re-emerging of SADS-CoV infection in pig herds in Southern China. Transbound Emerg Dis. 66(5):2180–2183.
- Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, Zhu Y, Zhang YW, Xie QM, Mani S, et al. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. 556(7700):255–258.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. 2020a. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579(7798):270–273.
- Zhou Z, Sun Y, Yan X, Tang X, Li Q, Tan Y, Lan T, Ma J. 2020b. Swine acute diarrhea syndrome coronavirus (SADS-CoV) antagonizes interferon-β production via blocking IPS-1 and RIG-I. Virus Res. 278:197843.