Abstract
Background: Macrophages constitute the main part of infiltrating immune cells in Malignant pleural mesothelioma (MPM) and abnormally high ratios of M2 macrophages are present in both pleural effusion and tissue samples of MPM patients. Whether MPM cells affect formation of M2 macrophages is poorly understood. In this study, we focused on identification of MPM-cells-derived soluble factors with M2-promoting effects. Methods: Media of malignant pleural mesothelioma cells were collected and soluble factors affecting macrophages were analyzed by mass spectrometry. TGF-β receptor inhibitor SB431542 was used as the entry point to explore the downstream mechanism of action by qRT-PCR, WB and immunofluorescence. Results: The serum-free culture media collected from the human MPM cells Meso1 and Meso2 significantly enhanced expression of the M2 signature molecules including IL-10, TGF-β and CD206 in the human macrophages THP-1, while the culture medium of the human MPM cells H2452 did not show such M2-promoting effects. Analysis of proteins by mass spectrometry and ELISA suggested that Leucine rich α2 glycoprotein 1(LRG1) was a potential candidate. LRG1 time- and dose-dependently increased expression of the M2 signature molecules, confirming its M2-promoting effects. Furthermore, LRG1’s M2-promoting effects were reduced by the TGF-β receptor inhibitor SB431542, and LRG1 increased phosphorylation of Smad2, indicating that M2-promoting effects of LRG1 were via the TGF-β receptor/Smad2 signaling pathway. Conclusions: Our results provide a potential M2-promoting new member, LRG1, which contributes to the immune escape of MPM via the TGF-β receptor/Smad2 signaling pathway.
Introduction
Malignant pleural mesothelioma (MPM), a malignancy developing from the mesothelium lining on the pleural cavity, is one of the most aggressive cancers. The etiology of MPM has been proven to be intimately associated with occupational asbestos exposure, characterized by a very long latency period of 30–40 years.Citation1,Citation2 Asbestos-induced carcinogenesis of mesothelial cells is a very long and complex process with inflammatory, molecular, genetic and epigenetic alterations.Citation3 Inhaled asbestos fibers can advance deep into the lung and reach the pleura, causing repeated cycles of irritation, inflammation and repair in the fiber-deposited sites.Citation4 The fiber-induced reactive oxygen species promote DNA damage and malignant transformation, while inflammatory cytokines and growth factors released from inflammatory cells, mesothelial cells and other cells accelerate inflammation and tumor growth.Citation5,Citation6
Macrophages are multi-functional innate immune cells with phagocytosis, cytokine production, antigen presentation and other functions. Due to their plasticity, macrophages can be activated to two functionally different phenotypes schematically designated as M1 and M2. Both phenotypes play critical roles in the asbestos-induced pathogenesis. As scavengers, resident alveolar macrophages clear inhaled asbestos fibers by phagocytosis.Citation7 Upon activation to the M1 phenotype by the fibers, they produce a variety of pro-inflammatory cytokines and chemokines such as IL-1β, TNF-α and MIP-1α to elicit inflammatory responses and attract monocytes and other immune cells to the fiber-deposited sites, resulting in local inflammatory lesions and fibrosis.Citation8,Citation9 When asbestos-induced lung cancer and MPM have formed, parts of tumor-associated macrophages (TAMs) become activated to the M2 phenotype. This moment they inhibit anti-tumor immune responses by secreting IL-10 and TGF-β, thus helping tumor cells growth and escape from immune surveillance.Citation10
Macrophages have been demonstrated to constitute the main part of infiltrating immune cells in MPM,Citation11 and abnormally high ratios of M2 macrophages are present in both pleural effusion and tissue samples of MPM patients.Citation12 Although many studies have demonstrated that formation of M2 macrophages in the tumor microenvironment (TME) is induced by IL-4, IL-13, IL-10, TGF-β and other factors through several signaling pathways in other cancers,Citation13,Citation14 the mechanisms of M2 macrophage induction are poorly understood. In this study, we focused on identification of MPM cells-derived soluble factors with a promoting effect on M2 macrophage formation, and found that leucine-rich alpha-2-glycoprotein 1 (LRG1) was such a potential factor.
Materials and methods
Culture of mesothelial cells and concentration of the culture supernatants
1 × 106 immortalized human normal mesothelial cells Met5A or 3 lines of human malignant pleural mesothelioma cells (Meso1, Meso2 and H2452; Meso1 and H2452 are epithelioid, Meso2 is sarcomatoid) were inoculated in 100 mm culture dishes containing RPMI1640 medium supplemented with 10% fetal bovine serum (Biological Industries, Israel) and cultured at 37 °C, 5% CO2 until the cells reached approximately 50% confluence. The cells were washed with PBS for 3 times and continued to culture in serum-free X-VIVO 15 (Lonza, Belgium) until the cells reached approximately 90% confluence. After removal of cell debris by centrifugation at 4 °C, 1500 rpm for 5 min, the culture media were concentrated 10 folds by 15 mL Vivospin concentrators (5000 MWCO PES, Satorius, Germany). The concentrated supernatants were stored at −20 °C for later use.
Stimulation of THP-1 cells with the culture supernatants
1 × 106 human macrophage/monocyte THP-1 cells were implanted in 6-well culture dishes containing RPMI1640 medium supplemented with 10% fetal bovine serum (Biological Industries) and cultured overnight at 37 °C, 5% CO2. The cells were then stimulated with 50 ng/mL phorbol-12-myristate-13-acetate (PMA, Sigma Aldrich) for 12 h to allow initial activation and cell attachment, and the culture media were replaced by 2 mL of 9:1 mixtures of fresh X-VIVO 15 and the 10-fold concentrated supernatants from Met5A, Meso1, Meso2 or H2452. 24 h later, the culture media were collected for ELISA analysis of cytokines, and the cells were used for qPCR.
Reverse transcription and qPCR
Total RNA of the THP-1 cells stimulated with the culture supernatants of Met5A, Meso1, Meso2 or H2452 was isolated using the TRIzol reagent (Invitrogen), and 1 µg of the total RNA was reverse-transcribed by the HiScript® IIRT SuperMix kit (Vazyme Biotech, Nanjing, China). qPCR analysis of M2 (IL-10, TGF-β, CD206) and M1 (IL-1β, TNF-α and iNOS) signature molecules were performed by the AceQ® qPCR SYBR Green Master Mix (Vazyme) as following cycles: 95 °C for 3 min; 95 °C for 12 sec and 60 °C for 40 sec, 40 cycles. The gene-specific primers are listed in the Table S2 (Supplementary material). Relative expression of the genes was calculated as 2−ΔΔCT, using GAPDH as an internal reference.
Enzyme-linked immunosorbent assays
Detection of human IL-10, TGF-β, IL-1β, and TNF-α in the supernatants of the THP-1 cells stimulated with the culture supernatants of Met5A, Meso1, Meso2 or H2452 and detection of LRG1 in the serum-free X-VIVO 15 media of Met5A, Meso1, Meso2 and H2452 cells were performed with human ELISA kits (FineTest, Wuhan, China) according to the manufacturer’s instructions.
Immunofluorescence
For observation of CD206 expression in THP-1 cells, 1 × 105 THP-1 cells were implanted in an 8-well chamber slide and stimulated with 50 ng/mL PMA for initial activation and cell attachment as described above. The culture media were then replaced by 0.5 mL of 9:1 mixture of fresh X-VIVO 15 and the 10-fold concentrated supernatants from Met5A, Meso1, Meso2 or H2452, or replaced by 0.5 mL fresh X-VIVO 15 containing 16 ng/mL recombinant human LRG1 (R&D systems, Minneapolis). Twenty-four hours later, the attached cells were fixed with 4% paraformaldehyde for 24 h. After PBST washing, the cells were blocked in 5% BSA-containing PBS for 1 h at room temperature, and incubated with rabbit anti-CD206 monoclonal antibody (1:200, Cell Signaling) at 4 °C overnight. Washed with PBST, the cells were then incubated with FITC-conjugated goat anti-rabbit IgG for 1 h at room temperature. The side was finally mounted with DAPI-containing mounting solution (Vazyme). Images of the randomly selected fields (20×) were taken with a fluorescence microscope (ECLIPSE, Nikon, Japan).
SDS-PAGE and mass spectrometry
10 µL of the 10-fold concentrated supernatants from Met5A, Meso1, Meso2 or H2452 was subjected for 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The gel was silver-stained with a silver staining kit (BestBio, Shanghai, China) and then analyzed by Mass Spectrometry (Beijing Genomics Institution, Shenzhen, China).
Stimulation of THP-1 cells with LRG-1
1 × 106 THP-1 cells were implanted in 6-well culture dishes, cultured in 10% fetal bovine serum RPMI1640 medium overnight at 37 °C, 5% CO2, and then stimulated with 50 ng/mL PMA for 12 h to allow initial activation and cell attachment. For observation of a dose-dependent response, the THP-1 cells were cultured in 2 mL fresh X-VIVO 15 containing different dose (0, 2, 4, 8, 16 and 32 ng/mL) of recombinant human LRG1 (R&D systems, Minneapolis) for 24 h; For observation of a time-dependent response, the cells were cultured in 2 mL fresh X-VIVO 15 containing 16 ng/mL) of recombinant human LRG1 for different time (0, 12, 24 and 48 h). The culture media were collected for ELISA analysis of cytokines, and the cells were used for qPCR.
Western blot
For analysis of phosphorylation of Smad2 and Smad3, 1 × 106 THP-1 cells were cultured in 6-well culture dishes and stimulated with PMA for initial activation and cell attachment. The cells were stimulated 16 ng/mL of recombinant human LRG1 with or without 5 µm of the TGF-β receptor inhibitor SB-431542 (CSN Pharm, Chicago) in 2 mL fresh X-VIVO 15 for 24 h. While the culture media were collected for ELISA analysis of cytokines, the cells were lysed in 100 µL of RIPA buffer. After protein quantification with BCA (BestBio, Shanghai, China), 10 µg of protein in the lysed solutions was separated by 10% SDS-PAGE and transferred onto PVDF membranes, and immunoblotted with rabbit anti-Smad2, anti-pSmad2, anti-Smad3 or anti-pSmad3 antibodies (ZenBioScience, Chengdu, China) diluted 1:200. The blots were washed and incubated for 1 h with biotinylated goat anti-rabbit IgG antibodies (Beyotime Biotech, Hongzhou, China) and then visualized using ECL Western Blotting Detection Reagent (Amersham Biosciences). GAPDH detected with anti-GAPDH antibody (dilution 1:1000; Peprotech) was used as an internal control. The optical intensity of the bands was analyzed using Image-Pro Plus (Media Cybernetics, Maryland).
Statistical analysis
Statistical analysis was performed using the SSPS17 software. The statistical significance was analyzed by two-tailed Student’s t test and One-way ANOVA, P values of <0.05 were considered to be significant.
Results
Soluble factors derived from MPM cells enhanced macrophage M2 phenotypes
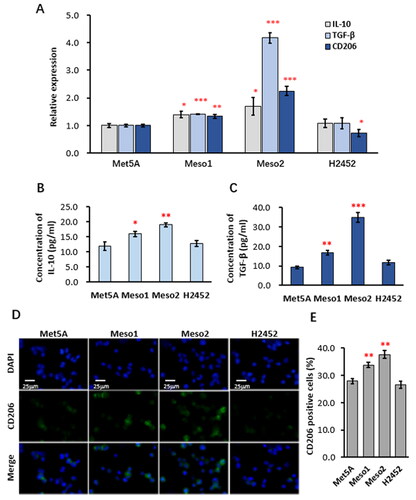
For observation whether soluble factors derived from MPM cells promote macrophage M2 polarization, the serum-free culture media were first collected from 3 lines of MPM cells (Meso1, Meso2 and H2452) and the normal mesothelial cells (Met5A). The culture media were then added to PMA-activated THP-1 cells, a human cell line of macrophage/monocyte. Detection by qRT-PCR revealed that the culture media of Meso1 and Meso2, but not H2452, significantly enhanced the expression of the macrophage M2 signature molecules IL-10, TGF-β and CD206, compared with that of Met5A (). Similarly, ELISA analyses showed that the concentrations of IL-10 and TGF-β in the culture medium of THP-1 cells were increased by treatment with the culture media of Meso1 and Meso2 (). Detection of CD206 expression by immunofluorescence () indicated that CD206-positive THP-1 cells in the Meso1 and Meso2 culture media-treated groups were 33.8 ± 1.0% and 37.7 ± 1.3%, respectively, significantly higher than that of Met5A (27.9 ± 1.0%), while the CD206-positive rate of the H2452 culture media-treated group (26.6 ± 1.3%) was comparable to that of Met5A (). Further examination of macrophage M1 signature molecules including IL-1β, TNF-α and iNOS by qRT-PCR (Supplemetary material, Figure S1A) and by ELISA (Supplemetary material, Figure S1B) revealed that the Meso1 and Meso2 culture media also suppressed the expression of these M1 signature molecules in the THP-1 cells. These results prove that some soluble factors derived from Meso1 and Meso2, but not from H2452, can enhance macrophage M2 phenotypes and suppress M1 phenotypes.
Figure 1. Soluble factors derived from Meso1 and Meso2 enhanced macrophage M2 phenotypes.
THP-1 cells were stimulated with 50 ng/mL PMA for 12 h to allow initial activation and cell attachment, and the culture media were replaced with 9:1 mixture of fresh X-VIVO 15 and the 10-fold concentrated supernatants from Met5A, Meso1, Meso2 or H2452. Twenty-four hours later, the culture media were collected for ELISA analysis of cytokines, and the cells were used for qPCR and immunofluorescence. (A) qPCR analysis of expression of the M2 signature molecules IL-10, TGF-β and CD206; (B) ELISA analysis of IL-10 and TGF-β; (C) Representative images of immunofluorescence staining of CD206-positive THP-1 cells; and (D) Percentage of CD206-positive THP-1 cells. Statistical analysis was from data of three independent experiments and the two-tailed Student’s t test was used for statistical significance. *, ** and ***, versus the Met5A group, indicate P values <0.05, 0.01 and 0.001, respectively.
LRG1 was identified as a potential candidate
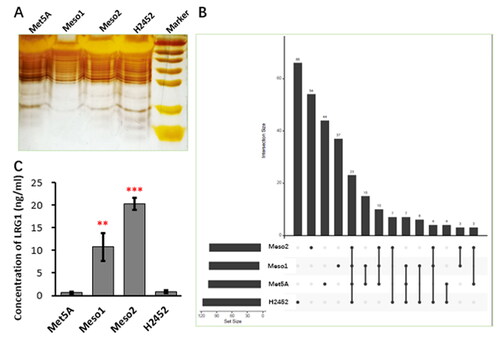
As described above, certain soluble factors derived from Meso1 and Meso2 had potential to induce macrophage M2 phenotypes. For analysis of what soluble factors were involved, 10-fold concentrated culture media from Met5A, Meso1, Meso2 and H2452 were undergone SDS-PAGE and silver-staining (), and the gel was subjected for mass spectrometry. As shown in the Venn graph ( and Supplemetary material, Table S1), total 282 proteins were identified, and only 3 proteins, Leucine-rich alpha-2-glycoprotein (LRG1), Polyamine-modulated factor 1-binding protein 1 (PMFBP1) and Switch-associated protein 70 (SWAP70) were only shared by Meso1 and Meso2. PMFBP1 and SWAP70 are not secretory proteins and possibly released from cells during cell turnover, and unlikely to mediate the observed M2 promotion effect of Meso1 and Meso2. LRG1 is a secretory protein involved in many pathogenic processes, and more likely to be the candidate. Examination of LRG1 by ELISA confirmed that only trace amount in the culture media of Met5A and H2452 was detected, while much higher content in the culture media of Meso1 and Meso2 was found ().
Figure 2. LRG1 was a potential candidate.
Ten-fold concentrated supernatants from Met5A, Meso1, Meso2 or H2452 were subjected for 10% SDS-PAGE, and the gel was silver-stained and then analyzed by Mass Spectrometry. (A) Image of the silver-stained gel; (B) The Venn graph of Mass Spectrometry results; (C) ELISA analysis of LRG1 in the 10-fold concentrated supernatants of Met5A, Meso1, Meso2, and H2452 cells. Statistical significance was analyzed from three independent experiments by the two-tailed Student’s t test. ** and *** indicate P values <0.01 and 0.001, respectively.
LRG1 dose- and time-dependently promoted macrophage M2 phenotypes
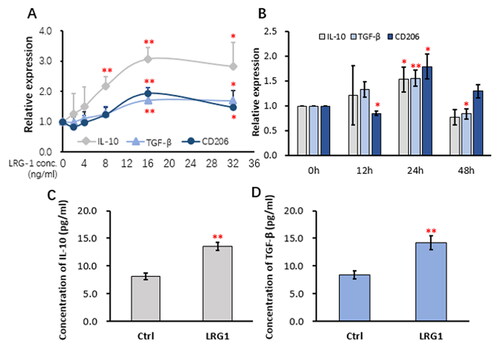
For understanding whether LRG1 stimulates dose-dependent responses, different doses of LRG1 (0, 2, 4, 8, 16 and 32 ng/mL) were given to the PMA-activated THP-1 cells for 24 h. As indicated in , qRT-PCR analysis showed that the mRNA expression of IL-10, TGF-β and CD206 was gradually increased with increasing LRG1 doses, reaching the peak at the dose of 16 ng/mL. Treatment of the PMA-activated THP-1 cells with 16 ng/mL of LRG1 for different time (0, 12, 24 and 48 h) induced increased mRNA expression of IL-10 and TGF-β, reaching the peak at 24 h (). An obvious time-dependent response for CD206 was not observed (). When the PMA-activated THP-1 cells were stimulated with 16 ng/mL of LRG1 for 24 h, the concentrations of IL-10 and TGF-β in the culture medium were elevated as detected by ELISA (). Taken together, LRG1 dose- and time-dependently enhances the expression of the M2 signature molecules IL-10 and TGF-β at both mRNA and protein levels. Checking the expression of M1 signature molecules by qRT-PCR found that while the expression of iNOS was decreased with increased LRG1 dosing, the expression of IL-1β and TNF-α was almost not affected by the LRG1 dosing (Supplemetary material, Figure S2A). Also, the expression of IL-1β was declined with increased time, but time-dependent expression of TNF-α and iNOS was not observed (Supplemetary material, Figure S2B). The concentration of IL-1β in the culture medium of the THP-1 cells treated with 16 ng/mL of LRG1 for 24 h was reduced as detected by ELISA, but the concentration of TNF-α was elevated (Supplemetary material, Figure S2C and D).
Figure 3. LRG1 elicited dose-and time-dependent responses in THP-1 cells.
THP-1 cells were stimulated with 50 ng/mL PMA for 12 h to allow initial activation and cell attachment. For observation of a dose-dependent response, the THP-1 cells were then cultured in fresh serum-free X-VIVO 15 containing different doses of recombinant human LRG1 for 24 h; For observation of a time-dependent response, the PMA-activated cells were cultured in fresh X-VIVO 15 containing 16 ng/mL of recombinant human LRG1 for different time. The culture media were collected for ELISA analysis of cytokines, and the cells were used for qPCR. (A) qRT-PCR analysis of the expression of IL-10, TGF-β and CD206 in THP-1 cells stimulated with different doses of LRG1 (0, 2, 4, 8, 16 and 32 ng/mL); (B) qRT-PCR analysis of the expression of IL-10, TGF-β and CD206 in THP-1 cells stimulated with 16 ng/mL LRG1 for different time (0, 12, 24 and 48 h); (C and D) ELISA detection of IL-10 and TGF-β in the supernatants of THP-1 cells stimulated with 16 ng/mL for 24 h. Statistical significance was analyzed from three independent experiments by One way ANOVA. * and **, versus the 0 ng/mL or 0 h groups, indicate P values <0.05 and 0.01, respectively.
Enhanced macrophage M2 phenotypes by LRG1 were via TGF-β receptors
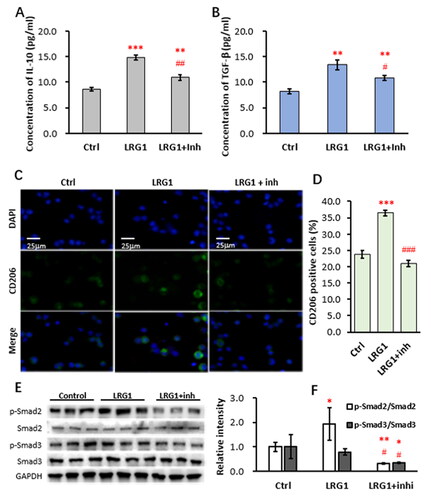
LRG1 is a member of TGF-β superfamily and associated with a variety of diseases and has been reported to enhance TGF-β signaling in kidney fibroblasts.Citation15,Citation16 Thus, we tested whether LRG1 promoted M2 phenotypes was via TGF-β receptors. As indicated in , stimulation of THP-1 cells with 16 ng/mL LRG1 for 24 h led to increased secretion of IL-10 and TGF-β into the culture media as detected by ELISA. When 5 µm of the TGF-β receptor inhibitor SB-431542 was added, the LRG1-enhanced production of IL-10 and TGF-β was recovered. Similarly, LRG1-induced decrease of IL-1β secretion and increase of TNF-α secretion were also restored by the inhibitor (Supplemetary material, Figure S3A and B). Further examination by immunofluorescence showed that LRG1-induced increase of CD206-positve THP-1 cells was also suppressed by the inhibitor (). LRG1 stimulation resulted in an increase of CD206-positve cells from 23.8 ± 1.1% to 36.4 ± 0.8%, while addition of the inhibitor restored the CD206-positve rate to 21.0 ± 1.1% (). Furthermore, Western blotting analysis of the downstream molecules Smad2 and Smad3 revealed that LRG1 did not change the expression of Smad2 and Smad3, but enhanced phosphorylation of Smad2, while phosphorylation of Smad3 was comparable (). Addition of the inhibitor SB-431542 led to marked decreases of phosphorylated Smad2 and Smad3 (). Relative intensities of phosphorylated Smad2 and Smad3 were shown in . These results suggest that LRG1 enhances macrophage M2 phenotypes possibly via the TGF-β receptor.
Figure 4. LRG1 enhanced M2 phenotypes via TGF-β receptor.
THP-1 cells were activated with 50 ng/mL PMA and then stimulated with 16 ng/mL of recombinant human LRG1 in the presence or absence of 5 µm of the TGF-β receptor inhibitor SB-431542 for 24 h. The culture media were used for ELISA detection and the cells were used for western blot and immunofluorescence. (A and B) ELISA detection of IL-10 and TGF-β in the supernatants of THP-1 cells; (C) Representative images of immunofluorescence staining of CD206-positive THP-1 cells; (D) Percentage of CD206-positive THP-1 cells; (E) western blot analysis of p-Smad2, Smad2, p-Smad3, and Smad3. GAPDH was used as an internal control; (F) The optical intensity of the bands analyzed using Image-Pro Plus. Statistical significance was analyzed from three independent experiments by the two-tailed Student’s t test. *, ** and ***indicate P values <0.05, 0.01 and 0.001; respectively, versus the control, while #, ## and ### indicate P values<0.05, 0.01 and 0.001; respectively, versus the LRG1 group.
Discussion
The immune microenvironment of MPM is very complicated, and infiltration of many types of immune cells, including macrophages, T lymphocytes, regulatory T cells, dendritic cells, natural killer cells, and innate lymphoid cells have been found in the MPM tissues.Citation17 However, most of the tumor-associated immune cells are functionally compromised, or have changed to forms that favor tumor growth and immune escape.Citation17 The high ratio of TAMs in the MPM tissues is possibly because MPM is a cancer induced by asbestos exposure. As phagocytes, alveolar macrophages and monocytes migrate to the sites where asbestos fibers have deposited, trying to clear the fiber by phagocytosis. NalP3 inflammasome-sensed phagocytosis of the fibers leads to secretion of pro-inflammatory cytokines and chemokines and recruitment of inflammatory cells.Citation18 Due to their undegradable nature, the asbestos fibers cannot be effectively cleared by macrophages, resulting in persistent inflammation. Failure of resolution of inflammation contributes to asbestos-induced carcinogenesis in the lung.Citation19 Nevertheless, how these pro-inflammatory M1 macrophages are initially triggered to change to immune-inhibitory M2 macrophages is still elusive.
LRG1 is a secretory protein, constitutively expressed by hepatocytes and neutrophils under physiological conditions, and increasingly expressed by endothelial cells, epithelial cells, fibroblasts and immune cells in response to many inflammatory stimuli.Citation20 Accumulating evidence indicates that LRG1 is involved in the pathogenesis of cancer, diabetes, inflammatory disorders and other diseases through the TGF-β canonical pathway and several other signaling pathway depending on different tissue contexts.Citation20 Immunologically, LRG1 is considered to an acute phase protein, as the serum level increases rapidly after microbial infection and other inflammatory stimuli,Citation21 suggesting that it is a component of the innate immune system. However, little is known about its effects on macrophages. LRG1 has been found to be required for the polarization and maintenance of specialized functions of peritoneal macrophages.Citation22 In the present study, we identified MPM cells-derived LRG1 as an M2 macrophage enhancing factor. LRG1 increased the expression of the M2 macrophage signature molecules including IL-10, TGF-β and CD206 via the TGF-β/Smad2. Although its M2 macrophage-promoting effects were week in our in vitro study, given that carcinogenesis of MPM is long term process, the effects might be accumulated in the development of MPM in the body. Also, LRG1-induced secretion of IL-10 and TGF-β can act on macrophages themselves in an autocrine manner, further enhancing M2 polarization of macrophages. It might serve as a switch to expedite M2 polarization of TAMs in the MPM tissue. LRG1 was found to secrete from only 2 cell lines in total 3 detected MPM cell lines, probably due to heterogeneity and genetic and epigenetic traits of MPM cells, suggesting that M2 macrophage-promoting mechanisms other than LRG1 may exist in the MPM microenvironment. Further studies concerning expression of LRG1 in the MPM tissue and its association with MPM development and formation of M2 macrophages are required.
In conclusion, LRG1 was identified to secrete from human mesothelioma cells and have an ability to enhance expression of the M2 signature molecules (IL-10, TGF-β and CD206) in the human macrophages THP-1. Inhibition of TGF-β receptor reduced the LRG1-induced expression of the M2 signature molecules and phosphorylation of Smad2, indicating that M2-promoting effects of LRG1 were via the TGF-β receptor/Smad2 signaling pathway. Our results provide a new member with potential M2-promoting effects and are helpful for understanding immune escape in MPM.
Author contributions
D.W. were responsible for cell culture, mass spectrometry, data analysis and the manuscript; W.P., Y.L. and R.M. for immunofluorescence and WB; W.G. and X.L. for qRT-PCR; Y.S. for ELISA; J.X., J.Y. and D.Z. for study design and manuscript revision.
-) supplemental_material_revised.docx
Download MS Word (156.5 KB)Declaration of interest
The authors declare that they have no conflict of interest.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Pairon J-C, Laurent F, Rinaldo M, et al. Pleural plaques and the risk of pleural mesothelioma. J Natl Cancer Inst. 2013;105(4):293–301. doi:10.1093/jnci/djs513.
- Wagner JC, Sleggs CA, Marchand P. Diffuse pleural mesothelioma and asbestos exposure in the north western cape province. Br J Ind Med. 1960;17(4):260–271. doi:10.1136/oem.17.4.260.
- Karpathiou G, Stefanou D, Froudarakis ME. Pleural neoplastic pathology. Respir Med. 2015;109(8):931–943. doi:10.1016/j.rmed.2015.05.014.
- Robinson BW, Musk AW, Lake RA. Malignant mesothelioma. Lancet. 2005;366(9483):397–408. doi:10.1016/S0140-6736(05)67025-0.
- Kamp DW, Israbian VA, Preusen SE, Zhang CX, Weitzman SA. Asbestos causes DNA strand breaks in cultured pulmonary epithelial cells: role of iron-catalyzed free radicals. Am J Physiol. 1995;268(3 Pt 1):L471–L480. doi:10.1152/ajplung.1995.268.3.L471.
- Sekido Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis. 2013;34(7):1413–1419. doi:10.1093/carcin/bgt166.
- Nishimura Y, Maeda M, Kumagai-Takei N, et al. Altered functions of alveolar macrophages and NK cells involved in asbestos-related diseases. Environ Health Prev Med. 2013;18(3):198–204. doi:10.1007/s12199-013-0333-y.
- Sanchez VC, Pietruska JR, Miselis NR, Hurt RH, Kane AB. Biopersistence and potential adverse health impacts of fibrous nanomaterials: what have we learned from asbestos? Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2009;1(5):511–529. doi:10.1002/wnan.41.
- Yang H, Bocchetta M, Kroczynska B, et al. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-κB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci USA. 2006;103(27):10397–10402. doi:10.1073/pnas.0604008103.
- Minnema-Luiting J, Vroman H, Aerts J, Cornelissen R. Heterogeneity in immune cell content in malignant pleural mesothelioma. Int J Mol Sci. 2018;19(4):1041. doi:10.3390/ijms19041041.
- Hegmans JP, Hemmes A, Hammad H, Boon L, Hoogsteden HC, Lambrecht BN. Mesothelioma environment comprises cytokines and T-regulatory cells that suppress immune responses. Eur Respir J. 2006;27(6):1086–1095. doi:10.1183/09031936.06.00135305.
- Salaroglio IC, Kopecka J, Napoli F, et al. Potential diagnostic and prognostic role of microenvironment in malignant pleural mesothelioma. J Thorac Oncol. 2019;14(8):1458–1471. doi:10.1016/j.jtho.2019.03.029.
- Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13(13):453–461. doi:10.2741/2692.
- Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci. 2021;22(13):6995. doi:10.3390/ijms22136995.
- Liu J-J, Pek SLT, Liu S, et al. Association of plasma leucine-rich alpha-2 glycoprotein 1 (LRG1) with all-cause and cause-specific mortality in individuals with type 2 diabetes. Clin Chem. 2021;67(12):1640–1649. doi:10.1093/clinchem/hvab172.
- Hong Q, Cai H, Zhang L, et al. Modulation of transforming growth factor-beta-induced kidney fibrosis by leucine-rich α-2 glycoprotein-1. Kidney Int. 2022;101(2):299–314. doi:10.1016/j.kint.2021.10.023.
- Désage A-L, Karpathiou G, Peoc’h M, Froudarakis ME. The immune microenvironment of malignant pleural mesothelioma: a literature review. Cancers (Basel). 2021;13(13):13. doi:10.3390/cancers13133205.
- Dostert C, Pétrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320(5876):674–677. doi:10.1126/science.1156995.
- Fishbein A, Hammock BD, Serhan CN, Panigrahy D. Carcinogenesis: failure of resolution of inflammation? Pharmacol Ther. 2021;218:107670. doi:10.1016/j.pharmthera.2020.107670.
- Camilli C, Hoeh AE, De Rossi G, Moss SE, Greenwood J. LRG1: an emerging player in disease pathogenesis. J Biomed Sci. 2022;29(1):6. doi:10.1186/s12929-022-00790-6.
- Shirai R, Hirano F, Ohkura N, Ikeda K, Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382(4):776–779. doi:10.1016/j.bbrc.2009.03.104.
- Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157(4):832–844. doi:10.1016/j.cell.2014.04.016.
