Abstract
Purpose and background: Paeoniflorin (PF) isolated from peony root (Paeoniae radix) has been used as a herbal medicine in East Asia for its anti-allergic, anti-inflammatory, and immunoregulatory effects. PF is known to cause apoptosis and to be a chemical heat shock protein (HSP) inducer. With this information, the effects on the gene expression in human leukemia U937 cells treated with PF were investigated.
Methods: U937 cells, a human myelomonocytic cell line, were treated with PF at different concentrations (0–640 µg/ml). Expression level of Hsp70 was monitored by Western blotting. Gene expression was evaluated using high-density oligonucleotide microarrays and computational gene expression analysis tools and the results were verified by real-time quantitative PCR.
Results: Although cell viability was not affected after PF treatment at a high concentration of 640 µg/ml, PF treatment (80–640 µg/ml) significantly elevated Hsp70 expression in a concentration-dependent manner. When the cells were treated with PF (160 µg/ml; 30 min), 35 up-regulated and 29 down-regulated genes were identified. Among the differentially expressed genes, a significant genetic network containing CDC2, FOSL1 and EGR1 was associated with biological functions such as cell death, gene expression or cellular growth and proliferation.
Conclusion: The present results indicate that PF affects the expression of many genes including Hsp70 and will provide a better understanding on the molecular mechanism of action of this compound in inducing HSPs in cells.
Introduction
Organisms respond to various physical, chemical and biological stresses by inducing the synthesis of highly conserved proteins called stress proteins or heat shock proteins (HSPs). The precise functions of HSPs are unknown, but considerable evidence points out that these stress proteins are essential for survival at both normal and elevated temperatures Citation[1]. Moreover, these rapidly synthesized HSPs function as molecular chaperones that act to regulate various cellular functions such as protein folding, refolding of partially denatured proteins, protein transport across membranes, cytoskeletal organizations, degradation of disabled proteins, and apoptosis, they also act as a cytoprotective factors against deleterious environmental stresses Citation[2–6]. Therefore, molecular chaperones are considered to be endogenous cytoprotective factors, lifeguards or guardians of proteom Citation[7], Citation[8]. Furthermore, recent studies have shown that HSPs placed in the extracellular environment activate target cells, particularly macrophages and antigen-presenting cells Citation[9]. HSPs are broadly categorized according to their molecular weight, which includes Hsp27, Hsp60, Hsp70, Hsp90 and Hsp100 families Citation[5]. Hsp70 is an important member of the HSP superfamily. It plays a key role in the process of protecting cells, facilitating the folding of nascent peptides and responding to stress Citation[10]. In addition to its function as an intracellular molecular chaperone, Hsp70 in the extracellular milieu acts as a powerful cytokine, affecting the functional properties of immunocompetent cells Citation[11].
Peony plants, such as Paeoniae suffruticosa, P. lactifora, P. veitchii, and P. obovata, have been used in traditional Chinese medicines or herbal medicines in China, Korea, and Japan. Paeoniflorin (PF), isolated from P. lactifora, is one of the major constituents of peony plants. Peony extracts and their constituents have been shown to have various biological and biomodulating activities, including improvement of memory Citation[12], antioxidant activity Citation[13], antiepileptic activity Citation[14], antimutagenic properties Citation[15], and antihyperglycemia Citation[16]. Although the molecular mechanism of these pharmacological functions of PF has not yet been elucidated, these activities might be ascribed in part to their positive effects on the induction of molecular chaperones like Hsp70 Citation[17].
Gene expression profiling is useful for clarifying changes at the molecular level occurring in experimental models. DNA microarray technology can provide a view of the expression profiles of hundreds or thousands of genes Citation[18], Citation[19]. Research papers applying DNA microarray technologies to analyse gene expression in a range of biological responses to physical and chemical stresses, such as heat shock Citation[20], ultrasound Citation[21], Citation[22] and bisphenol Citation[23], Citation[24] have been published from our laboratory.
In the present study, we investigated the optimal conditions in the induction of Hsp70 in human lymphoma U937 cells by PF. We also determined the gene expression profile of cells treated with PF with high-density oligonucleotide microarrays. To examine the ontology, including biological processes, cellular components, molecular functions and genetic networks, the data obtained were analysed using the Ingenuity Pathways Analysis tools and to verify the results of the microarray experiments, real-time quantitative PCR was performed.
Materials and methods
Chemical
PF (C23H28O11, MW: 480.46; Wako Pure Chemical Industries Ltd, Osaka, Japan) was dissolved in minimal amount of dimethyl sulfoxide (DMSO) and diluted with RPMI 1640 medium (Sigma-Aldrich, St Louis, MO, USA) to a final concentration of 80 µg/μl and stored in the freezer until use. The stock solution was diluted for use in fetal bovine serum (FBS) (Invitrogen, Tokyo, Japan) free culture medium.
Cell culture and treatments
U937 cells, a human myelomonocytic cell line, were obtained from the Human Sciences Research Resource Bank, Human Sciences Foundation (Tokyo, Japan). The cells were grown in RPMI 1640 medium supplemented with 10% FBS at 37°C. When the cells were treated with PF (0–640 µg/ml), the cells were incubated in RMPI 1640 medium (FBS-free solution) containing PF at desired concentrations for 30 min at 37°C. Final concentrations of DMSO used in the 80, 160, 240, 320, 400, 480, 560 and 640 µg/ml of PF were 0.02, 0.04, 0.06, 0.08, 0.10, 0.12, 0.14 and 0.16%. After treatment the cells were collected and suspended in fresh RPMI 1640 medium supplemented with 10% FBS and incubated at the desired times (3 h for dose-dependent experiments and 0–12 h in time-dependent experiments).
Measurements of cell viability and apoptosis
Cell viability of the cells was tested using Trypan blue exclusion test. Apoptosis was measured using DNA fragmentation assay Citation[25], Citation[26]. In short, the cells were lysed with a lysis buffer (1 mM EDTA, 0.2% Triton X-100 and 10 mM Tris-HCl, pH 7.5) and centrifuged at 13,000 × g for 10 min. Subsequently, each DNA in the supernatant and the pellet were precipitated in 12.5% trichloroacetic acid at 4°C and quantified using a diphenylamine reagent after hydrolysis in 5% trichloroacetic acid at 90°C for 20 min. The absorbance at 600 nm in each sample was determined after overnight colour development with diphenylamine reagent. The percentage of fragmented DNA in each sample was calculated as the amount of DNA in the supernatant divided by total DNA for that sample (supernatant plus pellet).
Western blotting
Western blotting was performed according to the previously described methods Citation[27], Citation[28]. Briefly, cells were collected at desired times, and washed in ice-cold phosphate buffered saline (PBS; 150 mM NaCl and 10 mM sodium phosphate, pH 7.4), then lysed with lysis buffer containing 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.05% sodium dodecyl sulfate (SDS) and 50 mM Tris-HCl (pH 7.4) on ice for 30 min. After a brief sonication (5 sec) the lysates were centrifuged at 13,000 rpm for 10 min at 4°C. The supernatants were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% gel and transferred to nitrocellulose membrane. After blotting with skimmed milk, the membrane was probed with monoclonal anti-Hsp70 (SR-B812; MBL, Nagoya, Japan) or anti-β-actin (AC-15, Sigma-Aldrich) antibody for 1–2 h at room temperature. After washing, the membrane was incubated with horseradish peroxidase-conjugated anti-mouse IgG for 1 h at room temperature. The membrane was then treated with enhanced chemiluminescence reagent (Amersham Biosciences, Buckinghamshire, UK), and the signals were detected by exposure of the membrane to X-ray films. Bands of target proteins were quantified by densitometry. Relative induction was normalized by β-actin.
Separation of total RNA
Total RNA was extracted from the cells using an RNeasy Total RNA Extraction Kit (Qiagen K.K., Tokyo, Japan). Then, RNA samples were treated with RNase-free DNase (Qiagen K.K.) for 30 min at room temperature.
Gene expression analysis
For gene expression analysis, we used a GeneChip® system with Human Expression Array U133A which was spotted with 22,283 probe sets (Affymetrix, Santa Clara, CA, USA). Sample preparation for array hybridization was carried out according to the manufacturer's instructions. Biotin-labelled cRNA probes were generated from 5 µg of DNase-treated total RNA. After fragmentation, the biotinylated cRNA probe was hybridized to arrays at 45°C for 16 h. The arrays were washed, stained with streptavidin-phycoerythrin and scanned with a probe array scanner. The scanned chip was analysed using the GeneChip Analysis Suite software (Affymetrix). Hybridization intensity data were converted into a presence/absence call for each gene, and changes in gene expression between experiments were detected by comparison analysis. The data were further analysed using GeneSpring software (Silicon Genetics, Redwood City, CA, USA) to extract the significant genes Citation[24].
Computational gene expression analysis
To examine the gene ontology, including biological processes, cellular components, molecular functions and genetic networks, the data were analysed using Ingenuity Pathways Analysis Tools (Ingenuity Systems, Mountain View, CA, USA), a web-delivered application that enables the discovery, visualization and exploration of molecular interaction networks in gene expression data. The gene lists identified by GeneSpring containing Affymetrix gene ID and natural legalism were uploaded into the Ingenuity Pathways Analysis Citation[24].
Real-time quantitative PCR assay
Real-time quantitative PCR was performed on a real-time PCR system (Mx3000P, Stratagene Japan K.K., Tokyo, Japan) using Brilliant SYBR Green qPCR Master Mix (Stratagene Japan K.K.) according to the manufacturer's protocol. Reverse transcriptase reaction was carried out with total RNA by using an oligo d(T)6 primer. Real-time quantitative PCR was performed by using the specific primers listed in . Each mRNA expression level was normalized with respect to the mRNA expression of glyceraldehydes-3-phosphate dehydrogenase (GAPDH) Citation[22].
Table I. Nucleotide sequences of primers for target genes.
Statistical analysis
Data are shown as means ± SD. Statistical analysis was carried out using Student's t test and P values less than 0.05 were regarded as significant.
Results
Effects of PF on U937 cells
Human leukemia U937 cells were treated with PF at different concentrations in FBS-free medium for 30 min. After treatment the cells were collected and suspended in fresh medium supplemented with 10% FBS and incubated at different times at 37°C. Cell viability was not affected after PF treatment at concentrations of 160 and 640 µg/ml (data not shown). Moreover, apoptosis was not observed even at a high concentration of 640 µg/ml. The percentage of DMSO used to dissolve PF was kept to the lowest level (0.04–0.16%) and had no effect on cell viability (data not shown).
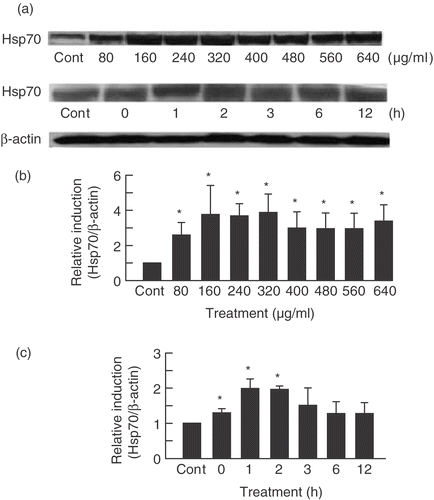
Expression level of Hsp70 in U937 cells treated with PF was monitored by Western blotting. Hsp70 expression was induced by PF treatment in a dose- and time-dependent manner (). Hsp70 was induced even at a relatively low concentration of PF (80 µg/ml) () and immediately after treatment (0 h incubation) (). DMSO at concentrations from 0.04 to 0.16% did not affect the expression of Hsp70. Considering the induction of Hsp70 by PF, on the basis of these data we chose 160 µg/ml for 30 min PF treatment, for the remainder of our studies.
Figure 1. PF induced Hsp70 in a dose- and time-dependent manner. (a) SDS-PAGE and Western blotting were performed on U937 cells treated with PF for 30 min at different concentrations (80–640 μg/ml for dose-dependent experiments and 160 µg/ml for time-dependent experiments). After which cells were incubated at different times (0–12 h for time-dependent experiments and 3 h for dose-dependent experiment). The signals were visualized by a luminescent image analyser using an ECL system. (b,c) Bands were quantified by densitometry and normalized with β-actin. Data are presented as mean ± SD (n = 3). *p < 0.05 vs. control (Student's t test). Cont, control.
Identification of genes responsive to PF treatment
To identify genes responsive to PF treatment in U937 cells, we carried out global-scale DNA microarray analysis of cells cultured at 0, 1, and 3 h after PF treatment (160 µg/ml, 30 min). Of approximately 22,300 genes analysed, 64 genes (35 up- and 29 down-regulated genes) that differentially expressed were identified in PF-treated cells in comparison with control cells. As shown in and , we classified the genes we identified as follows: (1) transiently increased genes, (2) gradually increased genes; (3) transiently decreased genes, and (4) gradually decreased genes. HSPs such as heat shock 70kDa protein 1A (HSPA1A) and 1B (HSPA1B) were included in the group of gradually increased genes. Transiently increased and gradually increased genes include FOS-like antigen 1 (FOSL1) or immediate early response 3 (IER3) and activating transcription factor 5 (ATF5) or chemokine (C–C motif) ligand 2 (CCL2), respectively (). Transiently decreased and gradually decreased genes include calreticulin (CALR) or interferon regulatory factor 2 (IRF2) and early growth response 1 (EGR1) or reticuloendotheliosis viral oncogene homolog (REL), respectively (). Complete lists of genes from all samples are available on the Gene Expression Ommnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc =GSE8228).
Table II. Upregulated genes under PF treatment.
Table III. Downregulated genes under PF treatment.
Computational gene expression analysis
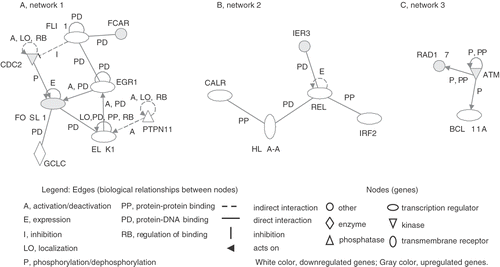
To examine the gene ontology, including biological processes, cellular components, molecular functions and genetic networks, the data identified here were analysed using the Ingenuity Pathway Analysis tools. Major biological functions of the differentially expressed genes influenced by PF were cell death, cancer and cell morphology. Among the differentially expressed genes, three significant genetic networks were identified. The genetic network 1 containing CDC2, FOSL1 and EGR1 was associated with cell death, gene expression or cellular growth and proliferation. Moreover, the genetic networks 2 and 3 containing REL and ATM were associated with immune response, cancer or cell death and cancer, cell cycle or DNA replication, recombination and repair, respectively ().
Figure 2. Genetic networks of genes associated with PF treatment. The cells were treated with PF (160 µg/ml, 30 min) and harvested after 3 h incubation. The genes associated with PF were analysed by the Ingenuity Pathway Analysis tool. The network is displayed graphically as nodes (genes) and edges (biological relationships between nodes).
Verification of differentially expressed genes
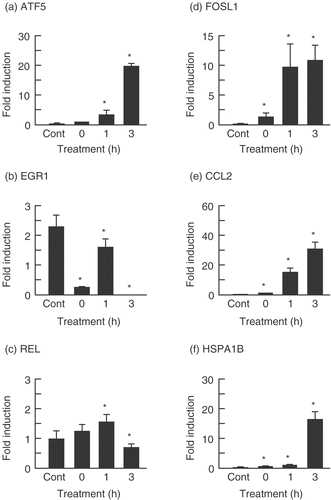
To verify the results of the microarray experiments, real-time quantitative PCR was performed. The results are summarized in . Although the expression levels of these genes were not completely comparable to that found by microarray analysis, the expression levels of ATF5, FOSL1, CCL2 and HSPA1B were significantly increased 3 h after PF treatment, with expression levels being, 21.5-, 9.4-, 31.5-, and 16.1-fold, respectively. In contrast, the expression levels of EGR1 and REL were significantly reduced, with expression levels of 0.007- and 0.63-fold, respectively.
Figure 3. Verification of microarray results with real-time quantitative PCR assay. The cells were treated with PF (160 µg/ml, 30 min) and harvested after incubation for 0, 1, and 3 h at 37°C. Each mRNA expression level was normalized with GAPDH. Data are presented as mean ± SD (n = 4). (a) ATF5 (Activating transcription factor (5); (b) EGR1 (Early growth response 1); (c) REL (Reticuloendotheliosis viral oncogene homolog); (d) FOSL1 (FOS-like antigen 1); (e) CCL2 (Chemokine (C–C motif) ligand 2); (f) HSPA1 (Heat shock 70 kDa protein B). *p < 0.05 versus control (Student's t test). Cont, control.
Discussion
HSPs have basic and indispensable functions in the life cycle of proteins as molecular chaperones Citation[6], as well as playing a role in protecting cells from environmental deleterious stresses Citation[2]. It is therefore beneficial to find substances that will induce HSPs without damaging the cells. Several researches have focused on natural products in search of HSP inducers Citation[17], Citation[29–31]. In the present study we demonstrated that treatment of human leukemia U937 cells with PF resulted in the induction of Hsp70 in a dose- and time-dependent manner. The concentration of PF used was much higher than previously reported but we treated the cells only for 30 min compared to 2 h of Yan's group Citation[17]. The cells were treated in FBS-free medium in this study as PF might bind to or be inactivated by serum proteins Citation[17]. Apoptosis was not observed because we used a much lower PF concentration and shorter time treatment compared to other studies Citation[32]. Induction of HSPs is mediated through the activation of a heat shock transcription factor (HSF1), which binds to conserved regulatory sequences called heat shock elements that are located in the promoter regions of inducible HSP genes. HSF1, which exists as inactive monomers in normal cells gets hyperphosphorylated, homotrimerizes and becomes transcriptionally competent upon stress Citation[33]. Yan et al. Citation[17] reported that the induction of HSPs by PF is mediated by the activation of HSF1. However, the molecular mechanism of this compound is still not completely understood.
With the high-density oligonucleotide microarrays and computational gene expression tools we identified a substantial number of genes with differential expressions between cells treated with PF and control cells. To our knowledge, this is the first report of DNA microarray analysis of genes that are differentially expressed in response to PF, a HSP-inducing compound. Of the 22,283 genes analysed, 35 genes were up-regulated while 29 genes were down-regulated. These data indicate that PF affects the expression of many genes including Hsp70. As expected, among the genes affected by PF treatment contains Hsp70 isoforms, HSPA1A and HSPA1B. In addition, increase in expression of HSPA1B was confirmed by real-time quantitative PCR. Previous report demonstrated that PF was able to induce proteins of Hsp27 and Hsp40 as well as Hsp70 in human cervical carcinoma HeLa cells and human neuroblastoma IMR-32 cells Citation[17]. In the current study, however, induction of mRNA levels of Hsp27 and Hsp40 in human leukemia U937 cells was not detected by using the GeneChip® Human Expression Array U133A which contained genes for these HSPs. This discrepancy may be due to different experimental conditions, including origin of the cell and exposure period and concentration of PF. Interestingly, we could identify three genetic networks which include differentially expressed genes elicited by PF treatment. Of these, the significant genetic network 1 containing CDC2, FOSL1 and EGR1 were associated with biological functions such as cell death, gene expression or cellular growth and proliferation. FOSL1 was reported to be up-regulated by stresses and its function is cellular defence response and positive regulation of cell proliferatio Citation[34], Citation[35]. It has also been indicated that CDC2 protein increases phosphorylation of FOSL1 protein in cell-free system Citation[36]. In the genetic network 3, interaction among RAD17, ATM and BCL11A were observed. It has been demonstrated that ATR/ATM-dependent phosphorylation of Rad17 is a critical early event during checkpoint signalling in DNA-damaged cells Citation[37]. Although the detailed mechanism of action of PF remains to be elucidated, the differentially expressed genes and genetic networks identified here are likely to be involved in HSP induction by this compound in cells.
Another gene of interest that was significantly increased after PF treatment is CCL2 which is involved in chemokine signalling. In a previous study, Hsp70 was found to stimulate cytokine production indicating that Hsp70 also protects cells by interacting with the innate and acquired immune system Citation[11]. In addition, Hsp70 has been shown to protect cells from necrosis, apoptosis and enable resistance of cells to various forms of stress and maintain cell survival Citation[38–40]. It therefore seems likely that Hsp70 and CCL2 may act as a protective molecule in the cell system.
In conclusion, the data presented here will provide a basis for further understanding of the molecular mechanisms of the role of Hsp70 induced by PF in cells.
Acknowledgement
This work was supported by a Grant-in-Aid for the 21st Century COE Program from the Ministry of Education, Culture, Sports, Sciences and Technology, Japan.
References
- Kregel KC. Heat shock proteins: Modifying factors in physiological stress responses and acquired thermotolerance. J Appl Physiol 2002; 92: 2177–2186
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Ann Rev Genet 1993; 27: 437–496
- Minami Y, Hohfeld J, Ohtsuka K, Hartl FU. Regulation of the heat shock protein 70 reaction cycle by the mammalian dnaj homolog, Hsp40. J Biol Chem 1996; 271: 19617–19624
- Michels AA, Kanon B, Konings AWT, Ohtsuka K, Bensaude O, Kampinga HH. Hsp70 and Hsp40 chaperone activities in the cytoplasm and the nucleus of mammalian cells. J Biol Chem 1997; 272: 33283–33289
- Di Maio A. Heat shock proteins. Facts, thoughts, and dreams. Shock 1999; 11: 1–12
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002; 295: 1852–1858
- Jaattela M. Heat shock proteins as cellular lifeguards. Ann Med 1999; 31: 261–271
- Ohtsuka K, Hata M. Molecular chaperone function of mammalian Hsp 70 and Hsp 40 - A review. Int J Hyperthermia 2000; 16: 231–245
- Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol 2002; 3: 185–194
- Arispe N, Doh M, Simakova O, Kurganov B, De Maio A. Hsc40 and Hsp70 interact with phosphatidylserine on the surface of PC12 cells resulting in a decrease of viability. FASEB J 2004; 18: 1636–1645
- Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. HSP70 stimulates cytokine production through a CD14-dependent pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med 2000; 6: 435–442
- Ohta H, Ni JW, Matsumoto K, Watanabe H, Shimizu M. Peony and its major constituents, paeoniflorin, improve radial maize performance impaired by scopolamine in rats. Pharmacol Biochem Behav 1993; 45: 719–723
- Okubo T, Nagai F, Seto T, Satoh K, Ushiyama K, Kano I. The inhibition of phenylhydroquinone-induced oxidative DNA cleavage by constituents of Moutan cortex and Paeoniae Radix. Biol Pharm Bull 2000; 23: 199–203
- Tsuda T, Sugaya A, Ohguchi H, Kishida N, Sugaya E. Protective effects of peony root extract and its components on neuron damage in the hippocampus induced by cobalt focus epilepsy model. Exp Neurol 1997; 146: 518–525
- Sakai Y, Nagase H, Ose Y, Kito H, Sato T, Kawai M, Mizuno M. Inhibitory action of peony root extract on the mutagenicity of benzo[a]pyrene. Mutat Res 1990; 244: 129–134
- Hsu FL, Lai CW, Cheng JT. Antihyperglycemic effects of paeoniflorin and 8-debenzoylpaeniflorin, glucosides from the root of Paeonia lactifora. Planta Med 1997; 63: 323–325
- Yan D, Saito K, Ohmi Y, Fujie N, Ohtsuka K. Paeoniflorin, a novel heat shock protein-inducing compound. Cell Stress Chaperones 2004; 9: 378–389
- DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, Chen Y, Su YA, Trent JM. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996; 14: 457–460
- Butte A. The use and analysis of microarray data. Nat Rev Drug Discov 2002; 1: 951–960
- Hirano H, Tabuchi Y, Kondo T, Zhao Q-L, Ogawa R, Cui Z-G, Feril LB Jr, Kanayama S. Analysis of gene expression in apoptosis in human lymphoma U937 cells induced by heat shock and the effects of α-phenyl N-tertbutylnitrone (PBN) and its derivatives. Apoptosis 2005; 10: 331–340
- Tabuchi Y, Kondo T, Ogawa R, Mori H. DNA microarray analyses of genes elicited by ultrasound in human U937 cells. Biochem Biophys Res Commun 2002; 290: 498–503
- Tabuchi Y, Ando H, Takasaki I, Feril LB Jr, Zhao QL, Ogawa R, Kudo N, Tachibana K, Kondo T. Identification of genes responsive to low intensity pulsed ultrasound in a human leukemia cell line Molt-4. Cancer Lett 2007; 246: 149–156
- Tabuchi Y, Kondo T. cDNA microarray analysis reveals chop-10 plays a key role in Sertoli cell injury induced by bisphenol A. Biochem Biophys Res Commun 2003; 305: 54–61
- Tabuchi Y, Takasaki I, Doi T, Ishii Y, Sakai H, Kondo T. Genetic networks responsive to sodium butyrate in colonic epithelial cells. FEBS Lett 2006; 580: 3035–3041
- Sellins KS, Cohen JJ. Gene induction by gamma-irradiation leads to DNA fragmentation in lymphocytes. J Immunol 1987; 139: 3199–3206
- Arai Y, Kondo T, Tanabe K, Zhao QL, Li FJ, Ogawa R, Li M, Kasuya M. Enhancement of hyperthermia-induced apoptosis by local anesthetics on human histiocytic lymphoma U937 cells. J Biol Chem 2002; 277: 18986–18993
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227: 680–685
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA 1979; 76: 4350–4354
- Ohtsuka K, Kawashima D, Gu Y, Kato K. Inducers and co-inducers of molecular chaperones. Int J Hyperthermia 2005; 21: 703–711
- Sõti C, Nagy E, Giricz Z, Vígh L, Csermely P, Ferdinandy P. Heat shock proteins as emerging therapeutic targets. Br J Pharmacol 2005; 146: 769–780
- Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J Biol Chem 2005; 280: 33097–33100
- Tsuboi H, Hossain K, Akhand AA. Paeoniflorin induces apoptosis of lymphocytes through a redox-linked mechanism. J Cell Biochem 2004; 93: 162–172
- Morimoto RI, Santoro MG. Stress-inducible responses and heat shock proteins: New pharmacologic targets for cytoprotection. Nat Biotechnol 1998; 16: 833–838
- Tsuchiya H, Fujii M, Niki T, Tokuhara M, Matsui M, Seiki M. Human T-cell leukemia virus type 1 Tax activates transcription of the human fra-1 gene through multiple cis elements responsive to transmembrane signals. J Virol 1993; 67: 7001–7007
- Zheng XH, Watts GS, Vaught S, Gandolfi AJ. Low-level arsenite induced gene expression in HEK293 cells. Toxicology 2003; 187: 39–48
- Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: Control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev 1998; 28: 370–490
- Bao S, Tibbetts RS, Brumbaugh KM, Fang Y, Richardson DA, Ali A, Chen SM, Abraham RT, Wang XF. ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 2001; 411: 969–974
- Yaglom JA, Ekhterae D, Gabai VL, Sherman MY. Regulation of necrosis of H9c2 myogenic cells upon transient energy deprivation. Rapid deenergization of mitochondria precedes necrosis and is controlled by reactive oxygen species, stress kinase JNK, HSP72 and ARC. J Biol Chem 2003; 278: 50483–50496
- Saleh A, Srinivasula SM, Balkir L, Robbins PD, Alnemri ES. Negative regulation of the Apaf-1 apoptosome by Hsp70. Nature Cell Biol 2000; 2: 476–483
- Steel R, Doherty JP, Buzzard K, Clemons N, Hawkins CJ, Anderson RL. Hsp72 inhibits apoptosis upstream of the mitochondria and not through interaction with Apaf-1. J Biol Chem 2004; 279: 51490–51499