Abstract
This paper compares methods for modeling the probability of removal when variable amounts of removal effort are present. A hierarchical modeling framework can produce estimates of animal abundance and detection from replicated removal counts taken at different locations in a region of interest. A common method of specifying variation in detection probabilities across locations or replicates is with a logistic model that incorporates relevant detection covariates. As an alternative to this logistic model, we propose using a catch–effort (CE) model to account for heterogeneity in detection when a measure of removal effort is available for each removal count. This method models the probability of detection as a nonlinear function of removal effort and a removal probability parameter that can vary spatially. Simulation results demonstrate that the CE model can effectively estimate abundance and removal probabilities when average removal rates are large but both the CE and logistic models tend to produce biased estimates as average removal rates decrease. We also found that the CE model fits better than logistic models when estimating wild turkey abundance using harvest and hunter counts collected by the Minnesota Department of Natural Resources during the spring turkey hunting season.
1. Introduction
The abundance of an animal population can often be estimated from counts that are modeled by a binomial or multinomial model with the exact model form dependent on the sampling method used to collect the counts. Royle and Dorazio Citation16 developed a hierarchical framework to model animal abundance and occurrence in a closed population from spatially replicated counts collected using a variety of sampling methods. One such sampling method, removal sampling, involves capturing and removing animals from a population over successive periods in time and possibly distinct spatial locations. Information about the population size just prior to the start of removal sampling is provided by the size and rate of decrease in successive removal counts.
Estimates of abundance based on a removal count model, or a similar replicated count model, are sensitive to the modeling assumptions made about the probability of removal, or detection, of animals in the population. Studies by Dorazio and Royle Citation3 Citation4 and Pledger Citation14, among others, showed that estimates of abundance can behave very poorly if the distribution used to model the probability of removal is not properly specified. Therefore, careful thought must be given about how to model the probability of removal and whether this probability is constant across sampling periods and sampling locations. Dorazio et al. Citation1 compared hierarchical models of abundance for analyzing removal counts obtained at spatially distinct locations and found that the best fitting models allowed site-specific removal probabilities. Such site-specific removal, or detection, probabilities can also be modeled with covariates that vary across sites or sampling periods. For example, Kéry et al. Citation11 and Forsyth et al. Citation6 used a logistic model for probability of detection that depended on sampling effort and habitat covariates.
This paper focuses on comparing removal probability models that incorporate a covariate that measures removal effort. We consider both a logistic regression specification for the removal probability and a catch–effort (CE) specification that assumes a Poisson process for removal effort [Chapter 7]Citation18. Both models will allow for heterogeneity in the removal probability by modeling it as a function of a measure of removal effort (e.g. number of hunters, number of lobster traps) for each sampling period and location. Logistic models of this type have been studied before but little research has been done on exploring the usefulness of a CE model for the removal probability in the hierarchical framework developed in Citation16. We applied both models to wild turkey harvest data collected by the Minnesota Department of Natural Resources (MN DNR) with the goal of determining which removal probability specification results in the best estimates of abundance and which is the most useful for predicting harvest rates for a given level of effort. We used Bayesian validation and goodness-of-fit (GOF) techniques to compare the fit of these models on the wild turkey harvest data and also conducted a simulation study to evaluate the behavior of each model for various population and removal scenarios.
Section 2 describes the logistic and CE models that will be compared and Section 3 outlines our Bayesian analysis and GOF tests. Section 4 describes the wild turkey and simulation results and Section 5 contains a discussion.
2. The model
2.1. Sampling method
Mark–recapture, point count, and removal surveys are common sampling methods used to estimate animal abundance or indices of abundance. In this paper, we will focus on the latter method of data collection and assume that removal counts were measured for at least two periods in time at spatially distinct locations. We will also include harvest counts collected during a hunting season in our definition of removal counts. This method of removing animals will induce a multinomial likelihood on the sequence of counts observed for each time period if we assume the population of animals in that region is closed, except for removals, during the removal (hunting) time periods. The regional data model is parameterized by N, the number of animals in the region, and p, the removal (harvest) probability for each time period. If more than one region is sampled, the full data likelihood will be the product of regional multinomial likelihoods which can model regional homogeneity (e.g. all regions might share the same p) or heterogeneity. We will consider models that allow for regional heterogeneity in both abundance and removal parameters.
Suppose m successive removal counts are observed in each of n distinct sample units surveyed. While these units could be points or line transects, we will assume that the sample units are non-overlapping areal regions since we are particularly interested in modeling harvest counts collected on this scale. Each region i contains a local population of N
i
animals prior to the start of the first removal period and during period j the removal probability p
ij
is the same for each animal in region i. Let y
ij
be the number of animals removed from region i during period j with giving the total number of animals removed in region i. The vector of removal counts
observed within a given region i has the following multinomial model:
2.2. Using removal effort to model the probability of removal
Let g ij measure the removal or hunting effort expended in region i during period j. We will consider two methods of modeling the removal probability p ij as a function of effort g ij .
2.2.1. A CE model
The first model for p
ij
that we will consider has a CE specification. Let θ
i
be the probability that an animal is removed with one unit of effort in region i. If we assume that effort is additive (i.e. each unit of effort acts independently and has the same probability of removing an animal) and θ
i
is constant across all n periods, then the probability of survival (non-removal) for an animal in region i during period j is . Our removal probability for an animal in this region and period is then
The per-unit removal probability θ i could be modeled in a variety of ways depending on the data available and assumptions made regarding the population and removal sampling. We will consider a simple fixed-effects model specification
Of course alternative models for θ
i
are possible. If no per-unit removal heterogeneity is present, we could use a uniform prior to model the constant removal probability . If the probability of removal could vary across regions and removal periods and relevant covariates are available, then we can model η
ij
as a function of time- and region-specific covariates x
ij
and parameters
which could also be region or time dependent.
2.2.2. A logistic regression model
The second type of specification for p ij that we will consider is a logistic regression model. We will then model the removal probability p ij as
2.2.3. Comparing models
displays removal probability p
ij
as a function of effort g
ij
for the CE and logistic specifications for four values of θ
i
and α1, i
. The intercept α0, i
was chosen so that p
ij
was equal to approximately 0.01 when effort was equal to 0 (). Under the CE specification, p
ij
will always be equal to 0 when effort is 0. The logistic model yields a sigmoidal function of effort for various values of α1, i
, while the CE model displays steep ascents to 1 for the larger values of θ
i
(0.10, 0.05) and an almost linear function of effort for small values of θ
i
(0.001). A more general comparison of the two model forms can be derived if we consider 1−p
ij
the probability that an animal survives (not removed during) a removal period. The logit model Equation(6)
assumes that each additional unit of effort will change the odds of survival by the factor
, while the CE model Equation(3)
assumes that each additional unit of effort will change the probability of survival by the factor 1−θ
i
(or
).
2.3. Modeling abundance
We will use a Poisson distribution to model local abundance N i
3. Bayesian analysis
The joint posterior for any of our proposed models will have the form
We used WinBUGS Citation12 and R Citation15 to fit and analyze the models described in Section 2 using Markov chain Monte Carlo (MCMC) simulations, but we preferred specifying a marginalized version of EquationEquation (10) since it had better MCMC convergence properties than did the full joint posterior described by EquationEquation (10)
. The marginalized model removed the local abundance parameters N
i
by summing the joint distribution of the data
and N
i
,
, over the values N
i
=y
i. to ∞ which induces a Poisson distribution with mean
on the individual removal counts y
ij
. After fitting this marginalized model, the posterior
can be approximated by using the data augmentation algorithm of Tanner and Wong Citation21 (see [pp. 286–290]Citation17 for more details on this marginalization and data augmentation process).
3.1. Model comparison
We considered several criteria when comparing our models. The deviance information criterion (DIC) suggested by Spiegelhalter et al. Citation20 measures both the fit and complexity of a model, with lower values given to good fitting, parsimonious models. DIC is the sum of p
D
, the effective number of parameters in the model, and , the posterior expectation of the deviance
. For the models described in Section 2, WinBUGS measures p
D
as
where
is the deviance measured at the posterior expectations of φ
i
and Ψ
i
.
We also computed the posterior predictive loss criterion suggested by Gelfand and Ghosh Citation7 as an alternative to DIC since work by Millar Citation13 suggests that DIC may not be a trustworthy comparison measure when fitting hierarchical models to count data. The posterior predictive criterion that minimizes the posterior predictive expectation under squared error loss is a linear combination of a GOF measure and a predictive variance measure
for an unknown replicate set of responses
. The predictive criterion is
for k>0 with smaller k resulting in larger penalties for models with larger predictive variance. Smaller values of D
k
indicate better fitting models. While there is no closed form of D
k
for our models, Monte Carlo estimates of G and P are easily obtained from the WinBUGS simulation results for each model.
3.2. Model validation
Models were also assessed using residuals and GOF measures computed for a validation sample that consisted of the removal counts that were observed during the last m−[mtilde] removal periods (1<[mtilde]<m) available in the data set. Let denote the set of responses that were used to fit a model and
denote the validation sample. The model under review was fit using
for all i=1, …, n regions and the validation set
was compared with the responses expected from the predictive posterior
. The joint distribution of
and
, given N
i
and Ψ
i
, is multinomial, which means that the posterior of
given
, N
i
and Ψ
i
is also a multinomial distribution with parameters adjusted appropriately. If we revert back to the original parameterization of the multinomial sampling model shown in EquationEquation (1)
, then this posterior is equal to
3.2.1. Residuals
The fit of the predictive posterior was analyzed by checking the Bayesian residuals of the validation sample. The Bayesian residuals were defined as
3.2.2. Goodness of fit
We also used the validation set to assess each model with a GOF test that used the χ2 discrepancy measure suggested by Gelman et al. [p. 175]Citation8. Three types of tests were considered, the first being an omnibus check of a model's ability to predict the number of removals made during the periods [mtilde]+1 through m. Since the validation sample is multinomial, the cumulative number of animals removed over the last m−[mtilde] removal periods, denoted as
, will have a binomial distribution with parameters Ñ
i
and
. The mean and variance for this binomial distribution will be used to form the first GOF test statistic:
As with any GOF test, we will compare the value of T
1 computed with the observed validation sample, denoted here as , to the values of T
1 that one would expected from totals [ttilde]
i
drawn from the binomial distribution specified above. A Bayesian p-value will be formed by weighting these binomial probabilities by the posterior distribution of the local abundance Ñ
i
and removal probabilities
used to compute the test statistic Equation(12)
. If
, then the Bayesian p-value for this GOF test is
(1) Generate a draw of | |||||
(2) Generate a realization of | |||||
(3) Compute the observed test statistic value | |||||
(4) Repeat steps 1 through 3 H times and [pcirc]
1 will be equal to the proportion of times that | |||||
The remaining two GOF tests are constructed in a similar manner. The second GOF test will assess how well a model can predict the number of removals made for each of the m−[mtilde] removal periods in the validation set. Recall that these individual removal counts are multinomial given , Ñ
i
and
, so the GOF test statistic will be
4. Examples
4.1. The wild turkey harvest
The wild turkey (Meleagris gallopavo) was successfully reintroduced in Minnesota during the 1960s and 1970s after its extirpation from the state in the late 1800s. In 1978, a spring turkey hunting season was started in southern Minnesota and by 2010 wild turkeys could be hunted legally throughout half of the state each spring during eight 5- to 7-day hunting periods. The wild turkey hunt is regulated by allocating a fixed number of hunting permits per season (hunting periods) for each turkey permit area (TPA) in the state.
An accurate assessment of the abundance of wild turkey populations in the state is needed for wildlife managers to determine permit allocations for each season. Demand for these permits is typically greater than the number of permits available, so deciding how many permits to allocate each year is important both for the viability of turkey populations and the satisfaction and safety of hunters. The MN DNR regulates the number of permits issued each year and requires hunters to register each harvested bird (only one bird per permit is allowed). Each recorded harvest indicates the location (TPA) and time period that the harvest occurred. We are interested in using hunter and harvest counts from the spring season to estimate the abundance of wild turkeys prior to the start of the season.
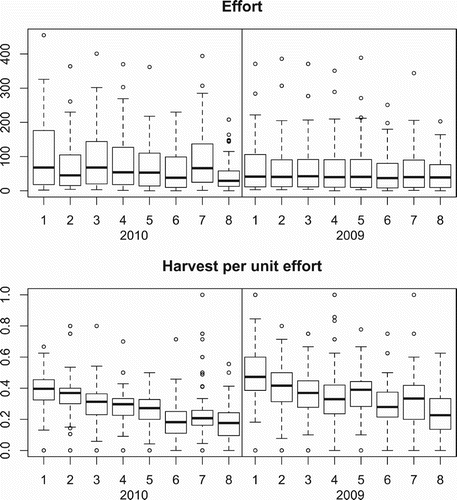
Our sample units are the TPAs open for hunting during the m=8 spring time periods. In 2010, there were n 10=77 TPAs and in 2009, there were n 09=76. The size A i of each TPA is measured in square miles. Removal effort g ij is measured by the number of permits issued for each TPA and time period combination. Harvest (removal) counts y ij are also measured for each TPA and time period combination, and since the vast majority of harvested birds during the spring season are male (e.g. 99.5% in 2010), we will use these harvest counts to assess the abundance of the male portion of the wild turkey population. shows removal effort and harvest per unit effort for 2010 and 2009. The effort in 2010 is higher and more variable than 2009. The total number of permits issued was 46,451 (603.3 per TPA) in 2010 and 36,187 (476.1 per TPA) in 2009. Even after accounting for the additional TPA opened in 2010, the total number of permits issued increased by over 10,000 from 2009 to 2010. The decrease in uniformity of effort across the eight time periods from 2009 to 2010 was primarily due to changes in spring turkey hunting regulations. The MN DNR increased the number of permits made available to hunters for time periods 7 and 8 by 110% from 10,582 in 2009 to 22,250 in 2010 and of these permits available for the 2010 season, approximately 46% were issued to hunters Citation5. Both years show a decreasing trend in harvest per unit effort over the 8 time periods, which somewhat levels out during the final weeks of the season. This trend is not surprising since the removal of birds in early time periods will leave fewer available for harvest in later time periods.
Figure 2. Effort, measured by number of hunting permits issued, and harvest per unit effort for the eight hunting periods (1 through 8) in 2009 and 2010.
The models that we considered assumed that p
ij
was the probability of removal for each wild turkey and each of the g
ij
permit holders used the same amount of effort to attempt to harvest one bird during hunting period j in region i. We fit each model separately for 2009 and 2010 since our main goal in this paper is to assess the fit of an abundance models for a given year rather than tracking abundance trends over multiple years (which is future work). Section 2.2.1 describes the two CE models that we considered: the ‘CE’ model refers to the fixed-effects specification Equation(4) of the per-unit removal probability θ
i
and the ‘hierarchical catch–effort’ (HCE) model refers to the hierarchical specification of θ
i
given in EquationEquation (5)
. Estimates from these CE models were compared to estimates obtained from the two logistic models described in Section 2.2.2: the ‘Logit’ model refers to the fixed-effects specification of the removal probability p
ij
given in EquationEquation (7)
, while ‘HLogit’ refers to the hierarchical modeling of this parameter shown in EquationEquation (8)
.
Vague prior distributions were given to most model parameters. For the abundance model described in Section 2.3, we placed a normal prior with mean 0 and variance 100 on the meta-level mean μφ and a uniform prior from 0 to 5 on the meta-level standard deviation σφ. Identical priors were used for the meta-level mean μη and standard deviation ση parameters for the HCE model in EquationEquation (5). In the HLogit model Equation(8)
, N(0, 100) priors were chosen for
and uniform(0, 10) priors where used for the standard deviations
(k=0, 1). Priors for the fixed-effect CE and Logit models in EquationEquations (4)
and Equation(7)
were also normal distributions centered at a
i
=d
k, i
=0 with a variance of
for k=0, 1 and i=1, …, n.
WinBUGS was used to fit each model using three chains of length 100, 000 with a burn-in of size 50, 000 and a thinning frequency of 30, resulting in a final simulation size of 5000. Convergence was assessed by the scale reduction factor Citation9 and a visual inspection of the mixing of the chains created for each parameter. Most parameters had scale reduction factors within 0.01 of 1 and all factors were with 0.09 of 1, which, along with the visual inspection, indicated adequate convergence for all models.
compares the CE and logistic models using DIC and the posterior predictive loss criterion discussed in Section 3.1. The values of k=1 and k=9 put a downscaling factor of 0.50 and 0.90, respectively, on the GOF measure G when estimating D
k
. Interestingly, the DIC measure favors the CE models in both years while the posterior predictive loss criterion D
k
only favors these models in 2010. In 2009, both CE models have smaller posterior variance (P) but higher GOF measures (G) when compared with the logistic models. Thus, when k=1 the CE looks like the best model choice but when k=9 the HLogit does slightly better. The non-hierarchical parameterizations of both the CE and logistic models decreased the average deviance of the model when compared with the hierarchical version (similar results were shown in Citation1) but the complexity and predictive variances of the hierarchical models were smaller since they allowed regional removal probability parameters (either η
i
or β
k, i
) to be shrunk toward a common mean (either μη or ).
Table 1. Model comparison using DIC (with average deviance ¯Dev and the effective number of parameters p d ) and using the estimated posterior predictive loss criterion with k=1 and k=9.
Model validation was done using the first [mtilde]=4 time periods to fit each model and harvest counts from the last four time periods (periods 5–8) were used as the validation sample. Analysis of the Bayesian residuals (Section 3.2.1) for the validation sample shows that the 2010 standardized residuals were fairly similar for both CE and logistic models but there was slightly less variation (extreme values) for the CE models for the 2009 validation sample. In general, both models seem slightly more prone to overestimation of the harvest counts during the validation sample time periods. This overestimation suggests that there may be some TPA where the removal probability decreases at some point during the hunting season.
The first GOF test indicates no significant lack-of-fit in each model's ability to predict the total harvest for time periods 5–8 (CE: [pcirc]
1 equals 0.44 and 0.42 for 2009 and 2010, respectively; HCE: 0.61 and 0.49; Logit: 0.42 and 0.22; HLogit: 0.56 and 0.25). All models do show a significant lack-of-fit in at least one TPA when predicting the time-period-specific harvest counts (all ). With 76 and 77 individual GOF tests (test 3) for each year, we would expect about 4 of the TPA-level tests to be significant at the 5% level if no lack-of-fit was present. The CE models for both years have between 16 and 18 regions with [pcirc]
3 less than 5%, while the logistics models for both years have between 15 and 33 significant TPA-level tests at the 5% level. displays which TPAs have significant lack-of-fit at the 5% and 1% levels, with regions in the southeastern portion of Minnesota standing out for all models in both 2009 and 2010 and regions in the northern range of the hunting region standing out in 2010. The lack-of-fit in these clusters of TPAs is likely due to violations in the assumptions of equal and independent per-hunter effort and constant per-hunter removal probability that all models share. The results of GOF tests suggest that there are some issues with GOF for some TPAs, but the tests also indicate that the CE models have better overall fit for both 2009 and 2010 when compared to the logistic models. Similar results were indicated by the model comparison criteria given in .
Figure 3. p-Values for individual TPA-level GOF tests (T 3) for 2009 (a) and 2010 (b). TPAs with GOF p-values [pcirc] 3 between 1% and 5% are shaded medium gray, while TPAs with p-values below 1% are shaded in dark gray. Regions with no significant lack-of-fit at the 5% level are shaded light gray.
![Figure 3. p-Values for individual TPA-level GOF tests (T 3) for 2009 (a) and 2010 (b). TPAs with GOF p-values [pcirc] 3 between 1% and 5% are shaded medium gray, while TPAs with p-values below 1% are shaded in dark gray. Regions with no significant lack-of-fit at the 5% level are shaded light gray.](/cms/asset/011e4969-2d45-4bb0-ace0-8e3406a073e5/cjas_a_748016_o_f0003g.jpg)
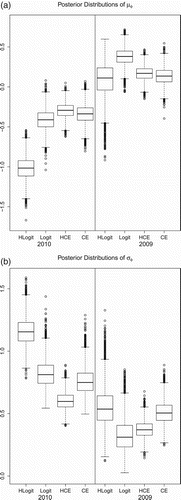
displays the posterior distributions for the meta-population level log-density parameters μφ and σφ. As indicated by the predictive posterior criteria [Pcirc] given in , the posterior distributions under the HCE model have less variation than the other models under consideration. Both CE models have similar posterior expectations for μφ but the posterior under the HLogit model supports smaller values of μφ than does the Logit model and both CE models. All models, expect the 2009 Logit model, support heterogeneity in log-density with posteriors for σφ that are not concentrated near 0.
gives estimates of the meta-population density parameters γ for the HCE and HLogit models. The mean and variance for the log-normal distribution of γ were computed as functions of the mean and standard deviation of where
and
. Monte Carlo standard errors were computed for each posterior mean estimate using the consistent batch means standard error estimator described in Citation10. Both models estimate mean regional density to be approximately 1.3 turkeys per square mile in 2009, while in 2010 the HCE model estimate drops to 0.90 and the HLogit model estimate drops to 0.72. The HCE model produces smaller credible interval for these parameters than the HLogit model. The HCE model also produces smaller estimates of
than the HLogit model for both years, most notably in 2010. This 2010 HLogit heterogeneity in the γ
i
is possibly due to the influence of the removal probability intercept parameter α0 which gives the logit-scaled removal probability at 0 effort. In 2010, the mean removal probability of HLogit with 0 effort is
for one period which results in unrealistically large harvest probabilities for TPAs that have small amounts of effort for each removal period. The overall harvest rate in these regions is larger for the HLogit model than all other models which results in density estimates that are much smaller than other models. In 2009, the mean removal probability with 0 effort for the HLogit model is much smaller with
. Note that we did explore logistic models with no intercept term but their fit was much poorer than the models that did include α0.
Table 2. Estimates of the metapopulation parameters for abundance and removal probability in the HCE and logit (HLogit) models.
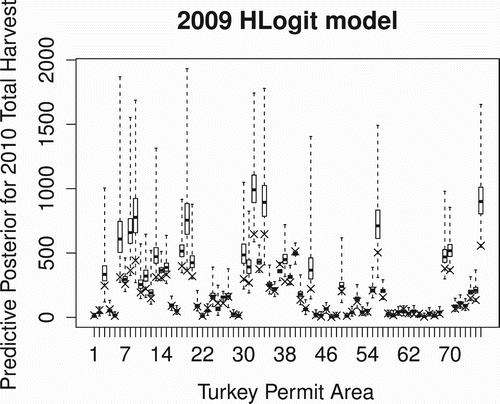
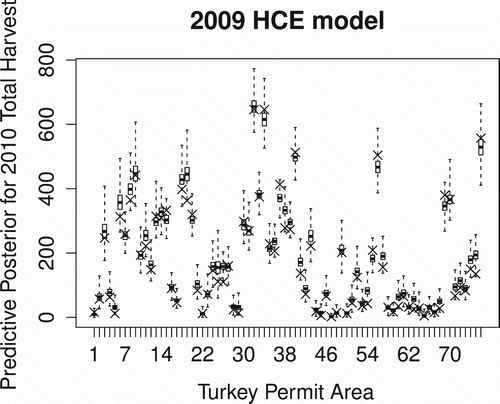
We also considered the management task of allocating hunting permits for an upcoming hunting season by assuming no change in abundance and removal probability so that we could use the 2009 models to estimate 2010 harvest totals for a given amount of effort. Using the actual number of hunters in 2010, we used the 2009 model to compute the predictive posterior distributions for the (unseen) 2010 harvest total for each of the 76 TPAs where hunting was permitted in 2009. and show these predictive posteriors for the HLogit and HCE models, respectively, along with an × marking the actual harvest total from 2010. While both models tend to overestimate the harvest total, likely due to the larger 2009 abundance estimates (), the HCE model would be the preferred model for prediction purposes since it produces predictive posteriors with smaller variation than the HLogit model.
4.2. Simulation results
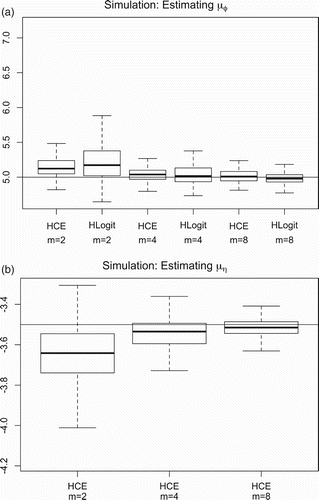
We also conducted a simulation to study properties of the HCE model proposed in this paper using removal counts generated from the catch-per-unit-effort process described in Section 2.2.1. For comparison purposes, we also fit the hierarchical logistic regression model (HLogit) in EquationEquation (8) to these simulated removal counts. The number of regions (n=30) and log-density parameters
were held fixed for the simulation, but the logit-scaled removal probability parameters
and number of removal periods (m) were allowed to vary. We also assumed that each region had equal area of A
i
=1 so that the mean density of about
also measures the mean abundance in each region. Removal effort varied throughout the regions and was designed to decrease from removal period 1 to m. For each unique data generating process, we obtained 100 realized removal counts and used WinBUGS to fit the HCE and HLogit models. The prior distributions and chain lengths described in Section 4.1 were also used in this simulation study. Parameter estimates (posterior means) and 95% credible intervals were obtained for each model and we computed the root mean square error (RMSE) and relative bias (
) for each estimator and the average credible interval length and coverage rate.
compares log-density parameter estimates and provides HCE removal probability estimates for data generated using m=8 removal periods and varying levels of hunter effort and catchability. The average total removal effort per region () was either 48.8 or 488 units of effort and the mean logit-scaled per-unit removal probability ranged from
to μη=−7. For each level of effort and mean removal probability, the average probability of harvest across all regions for the data generating process was calculated as
Table 3. Simulation results based on 100 sets of removal counts generated from the catch-per-unit-effort process described in Section 2.2.1.
shows simulation results for varying the number of hunting periods (m=2, 4, 8). The HCE and HLogit models show a tendency to overestimate the mean abundance μφ as the number of removal periods m decrease and the HCE model tends to underestimate μη as m decreases. We also considered a scenario with m=8 time periods and μφ=5 with μη equal to −3.5 during periods 1–4 and then dropped μη down to −3.7 during periods 5–8. We found that both the HLogit and HCE models underestimated abundance (relative bias of −3.1% and −1.7%, respectively) and have less than optimal 95% confidence interval coverage rates (68% and 84%, respectively). We were surprised to find that the HCE model produced an average estimate of that was greater than both of the values of μη used in the data generating process (−3.5 and −3.7). We think that this may have occurred because the HCE model overcompensated for the larger than expected drop in removal counts during periods 5–8 by overestimating the overall probability of removal (i.e. if more animals were harvested in the initial periods then there will be fewer to harvest in the later periods). This positive bias in estimating the removal probability is the reason for the negative bias in abundance estimates.
5. Discussion
The CE models for the wild turkey harvest data generally fit as good or better than the logistic removal probability models that we considered (). The validation sample GOF results indicate that all models can estimate total harvest for the last four periods given results from the first four periods (test 1), but all models show some lack-of-fit when broken down into an assessment of the per-period harvest count predictive capabilities. Again though, the CE models show less lack-of-fit than the logistic model ().
These results suggest that the catch-per-unit-effort specification for the per-period removal probability is preferred over the logistic model for our wild turkey harvest data. While the assumption of additivity in the CE model is strong, it is actually not any stronger than the assumptions inherent in the logistic model when using removal effort as a covariate. In the logit model, each additional unit of effort will change the odds of survival by the factor while with the CE model, each additional unit of effort will change the probability of survival by the factor 1−θ
i
. Regardless of the model, each additional unit of effort has the same effect on either the odds or probability of survival and violations of the additivity assumption will result in biased estimates from either model. Both models also currently assume that the effect of each unit of effort on the probability of removal (e.g. the parameter θ
i
or α1, i
) is the same across all eight hunting periods and the removal probability p
ij
is constant during hunting period j. As described by Seber Citation18, these assumptions are often not completely satisfied when using removal effort to estimate abundance. Weather conditions and variation in individual hunter ability are likely sources of day-to-day variation in the removal probability for each unit of effort. We could also see a small decrease in p
ij
during hunting period j due to the catch limit of one bird per permit which may result in a small decrease in effort during the 5–7-day-long hunting period. Our simulation results suggest that both models are prone to underestimating abundance when the removal probability drops halfway through an eight-period-long hunting season.
While the CE model we considered in this paper looks promising, we believe it could be improved by pursuing two avenues. First, we could relax the i.i.d. assumptions made for the TPA-level density γ i and per-unit removal probabilities θ i and allow some form of dependence to exist between TPAs at this level. One option would be to model densities using a conditionally autoregressive (CAR) model for one (or both) of these parameters as was done with point-count data by Webster et al. Citation22. Alternatively, Dorazio et al. Citation2 propose using a Dirichlet process prior to model φ i when we lack the covariates needed to explain variation in abundance across the region of study. While the CAR model will place a spatial dependence on the parameters, the Dirichlet process prior will allow a weaker dependence to exist between regions that may, for example, have similar densities due to similar habitat but which may not be located next to one another. The second avenue to pursue will be to add a temporal scale to the model so that we can estimate changes in abundance across multiple years. While the current (non-dynamic) version of the HCE model shows promise when predicting future harvest totals (), such predictions will likely be improved by allowing future abundance level to change over time with a dynamic population model. A temporal HCE model will be valuable from a management perspective since it will monitor abundance over multiple years and predict next years harvest rates based on any number of permit allocation (effort) scenarios.
Acknowledgements
This work was supported in part by a Hewlett Mellon Fellowship from Carleton College. The authors thank Dr John Fieberg, MN DNR, and Dr Robert Dorazio, US Geological Survey, for their suggestions on this paper.
Additional information
Notes on contributors
Eric Dunton
Present address: Shiawassee National Wildlife Refuge, US Fish, Wildlife Service, Saginaw, MI 48601, USA.References
- Dorazio , R. M. , Jelks , H. L. and Jordan , F. 2005 . Improving removal-based estimates of abundance by sampling a population of spatially distinct subpopulations . Biometrics , 61 : 1093 – 1101 . (doi:10.1111/j.1541-0420.2005.00360.x)
- Dorazio , R. M. , Mukherjee , B. , Zhang , L. , Ghosh , M. , Jelks , H. L. and Jordan , F. 2008 . Modeling unobserved sources of heterogeneity in animal abundance using a Dirichlet process prior . Biometrics , 64 : 635 – 644 . (doi:10.1111/j.1541-0420.2007.00873.x)
- Dorazio , R. M. and Royle , J. A. 2003 . Mixture models for estimating the size of a closed population when capture rates vary among individuals . Biometrics , 59 : 351 – 364 . (doi:10.1111/1541-0420.00042)
- Dorazio , R. M. and Royle , J. A. 2005 . Rejoinder to ‘The performance of mixture models in heterogeneous closed population capture-recapture’ . Biometrics , 61 : 874 – 876 . (doi:10.1111/j.1541-020X.2005.00411_2.x)
- Dunton , E. 2010 . Spring 2010 wild Turkey harvest report Agency Report, Minnesota Department of Natural Resources, St. Paul, MN
- Forsyth , D. M. , Link , W. A. , Webster , R. , Nugent , G. and Warburton , B. 2005 . Nonlinearity and seasonal bias in an index of brushtail possum abundance . J. Wildlife Manag. , 69 : 976 – 984 . (doi:10.2193/0022-541X(2005)069[0976:NASBIA]2.0.CO;2)
- Gelfand , A. E. and Ghosh , S. K. 1998 . Model choice: A minimum posterior predictive loss approach . Biometrika , 85 : 1 – 11 . (doi:10.1093/biomet/85.1.1)
- Gelman , A. , Carlin , J. , Stern , H. and Rubin , D. B. 2004 . Bayesian Data Analysis , 2 , Boca Raton , FL : Chapman & Hall/CRC .
- Gelman , A. and Rubin , D. B. 1992 . Inference from iterative simulation using multiple sequences . Statist. Sci. , 7 : 457 – 472 . (doi:10.1214/ss/1177011136)
- Jones , G. L. , Haran , M. , Caffo , B. S. and Neath , R. 2006 . Fixed-width output analysis for Markov chain Monte Carlo . J. Amer. Statist. Assoc. , 101 : 1537 – 1547 . (doi:10.1198/016214506000000492)
- Kéry , M. , Royle , J. A. and Schmid , H. 2005 . Modeling avian abundance from replicated counts using binomial mixture models . Ecol. Appl. , 15 : 1450 – 1461 . (doi:10.1890/04-1120)
- Lunn , D. J. , Thomas , A. , Best , N. and Spiegelhalter , D. 2000 . WinBUGS – a Bayesian modelling framework: Concepts, structure, and extensibility . Stat. Comput. , 10 : 325 – 337 . (doi:10.1023/A:1008929526011)
- Millar , R. B. 2009 . Comparison of hierarchical Bayesian models for overdispersed count data using DIC and Bayes’ factors . Biometrics , 65 : 962 – 969 . (doi:10.1111/j.1541-0420.2008.01162.x)
- Pledger , S. 2005 . The performance of mixture models in heterogeneous closed population capture–recapture . Biometrics , 61 : 868 – 876 . (doi:10.1111/j.1541-020X.2005.00411_1.x)
- R Development Core Team . 2011 . R: A Language and Environment for Statistical Computing , Vienna : R Foundation for Statistical Computing . Available at http://www.R-project.org (accessed June 30, 2011).
- Royle , J. A. and Dorazio , R. M. 2006 . Hierarchical models of animal abundance and occurrence . J. Agric. Biol. Environ. Stat. , 11 : 249 – 263 . (doi:10.1198/108571106X129153)
- Royle , J. A. and Dorazio , R. M. 2008 . Hierarchical Modeling and Inference in Ecology: The Analysis of Data from Populations, Metapopulations, and Communities , San Diego , , CA : Academic Press .
- Seber , G. A.F. 1982 . The Estimation of Animal Abundance and Related Parameters , 2 , London : Charles W. Griffin .
- Skalski , J. R. , Ryding , K. E. and Millspaugh , J. J. 2005 . Wildlife Demography: Analysis of Sex, Age, and Count Data , Burlington , MA : Elsevier Academic Press .
- Spiegelhalter , D. J. , Best , N. , Carlin , B. P. and van der Linde , A. 2002 . Bayesian measures for model complexity and fit (with discussion) . J. R. Statist. Soc. Ser. B , 64 : 583 – 639 . (doi:10.1111/1467-9868.00353)
- Tanner , M. A. and Wong , W. H. 1987 . The calculation of posterior distributions by data augmentation . J. Amer. Statist. Assoc. , 82 : 528 – 550 . (doi:10.1080/01621459.1987.10478458)
- Webster , R. A. , Pollock , K. H. and Simons , T. R. 2008 . Bayesian spatial modeling of data from avian point count surveys . J. Agric. Biol. Environ. Stat. , 13 : 121 – 139 . (doi:10.1198/108571108X311563)
![Figure 1. Probability of removal during one removal period for (a) the CE specification for θ i of 0.1, 0.05, 0.01, and 0.001 and (b) the logistic specification p ij =1/[1+exp(−α0, i −α1, i g ij )] with α0, i =−4.6 and α1, i of 0.1, 0.05, 0.01, and 0.001.](/cms/asset/b21cbb41-c3e8-402d-9e89-7cb8e0127a1f/cjas_a_748016_o_f0001g.gif)